A novel autosomal dominant GREB1L variant associated with non-syndromic hearing impairment in Ghana
- PMID: 36357908
- PMCID: PMC9648021
- DOI: 10.1186/s12920-022-01391-w
A novel autosomal dominant GREB1L variant associated with non-syndromic hearing impairment in Ghana
Abstract
Background: Childhood hearing impairment (HI) is genetically heterogeneous with many implicated genes, however, only a few of these genes are reported in African populations.
Methods: This study used exome and Sanger sequencing to resolve the possible genetic cause of non-syndromic HI in a Ghanaian family.
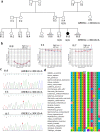
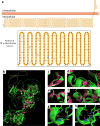
Results: We identified a novel variant c.3041G > A: p.(Gly1014Glu) in GREB1L (DFNA80) in the index case. The GREB1L: p.(Gly1014Glu) variant had a CADD score of 26.5 and was absent from human genomic databases such as TopMed and gnomAD. In silico homology protein modeling approaches displayed major structural differences between the wildtype and mutant proteins. Additionally, the variant was predicted to probably affect the secondary protein structure that may impact its function. Publicly available expression data shows a higher expression of Greb1L in the inner ear of mice during development and a reduced expression in adulthood, underscoring its importance in the development of the inner ear structures.
Conclusion: This report on an African individual supports the association of GREB1L variant with non-syndromic HI and extended the evidence of the implication of GREB1L variants in HI in diverse populations.
Keywords: GREB1L; Ghana; Hearing impairment.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Hereditary Hearing Loss Homepage. [https://hereditaryhearingloss.org/].
-
- Wonkam A, Bosch J, Noubiap JJ, Lebeko K, Makubalo N, Dandara C. No evidence for clinical utility in investigating the connexin genes GJB2, GJB6 and GJA1 in non-syndromic hearing loss in black Africans. S Afr Med J = Suid-Afrikaanse tydskrif vir geneeskunde. 2015;105(1):23–26. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

