Canine Saliva as a Possible Source of Antimicrobial Resistance Genes
- PMID: 36358144
- PMCID: PMC9686479
- DOI: 10.3390/antibiotics11111490
Canine Saliva as a Possible Source of Antimicrobial Resistance Genes
Abstract
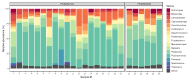
While the One Health issues of intensive animal farming are commonly discussed, keeping companion animals is less associated with the interspecies headway of antimicrobial resistance. With the constant advance in veterinary standards, antibiotics are regularly applied in companion animal medicine. Due to the close coexistence of dogs and humans, dog bites and other casual encounters with dog saliva (e.g., licking the owner) are common. According to our metagenome study, based on 26 new generation sequencing canine saliva datasets from 2020 and 2021 reposited in NCBI SRA by The 10,000 Dog Genome Consortium and the Broad Institute within Darwin's Ark project, canine saliva is rich in bacteria with predictably transferable antimicrobial resistance genes (ARGs). In the genome of potentially pathogenic Bacteroides, Capnocytophaga, Corynebacterium, Fusobacterium, Pasteurella, Porphyromonas, Staphylococcus and Streptococcus species, which are some of the most relevant bacteria in dog bite infections, ARGs against aminoglycosides, carbapenems, cephalosporins, glycylcyclines, lincosamides, macrolides, oxazolidinone, penams, phenicols, pleuromutilins, streptogramins, sulfonamides and tetracyclines could be identified. Several ARGs, including ones against amoxicillin-clavulanate, the most commonly applied antimicrobial agent for dog bites, were predicted to be potentially transferable based on their association with mobile genetic elements (e.g., plasmids, prophages and integrated mobile genetic elements). According to our findings, canine saliva may be a source of transfer for ARG-rich bacteria that can either colonize the human body or transport ARGs to the host bacteriota, and thus can be considered as a risk in the spread of antimicrobial resistance.
Keywords: antimicrobial resistance; bacteriome; dog saliva; mobilome; resistome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- European Centre for Disease Prevention and Control (ECDC) European Food Safety Authority (EFSA) European Medicines Agency (EMA) Third joint inter-agency report on integrated analysis of consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from humans and food-producing animals in the EU/EEA. EFSA J. 2021;19:e06712. doi: 10.2903/j.efsa.2021.6712. - DOI - PMC - PubMed
-
- Rowan A.N. Companion Animal Statistics in the USA. 2018. [(accessed on 6 August 2022)]. Available online: https://animalstudiesrepository.org/cgi/viewcontent.cgi?article=1002&con....
-
- Bedford E. Number of Dogs in the United States from 2000 to 2017. 2019. [(accessed on 6 March 2022)]. Available online: https://www.statista.com/statistics/198100/dogs-in-the-united-states-sin....
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

