Phage Cocktail in Combination with Kasugamycin as a Potential Treatment for Fire Blight Caused by Erwinia amylovora
- PMID: 36358221
- PMCID: PMC9686651
- DOI: 10.3390/antibiotics11111566
Phage Cocktail in Combination with Kasugamycin as a Potential Treatment for Fire Blight Caused by Erwinia amylovora
Abstract
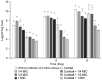
Recently, there has been an increasing number of blight disease reports associated with Erwinia amylovora and Erwinia pyrifoliae in South Korea. Current management protocols that have been conducted with antibiotics have faced resistance problems and the outbreak has not decreased. Because of this concern, the present study aimed to provide an alternative method to control the invasive fire blight outbreak in the nation using bacteriophages (phages) in combination with an antibiotic agent (kasugamycin). Among 54 phage isolates, we selected five phages, pEa_SNUABM_27, 31, 32, 47, and 48, based on their bacteriolytic efficacy. Although only phage pEa_SNUABM_27 showed host specificity for E. amylovora, all five phages presented complementary lytic potential that improved the host infectivity coverage of each phage All the phages in the cocktail solution could lyse phage-resistant strains. These strains had a decreased tolerance to the antibiotic kasugamycin, and a synergistic effect of phages and antibiotics was demonstrated both in vitro and on immature wound-infected apples. It is noteworthy that the antibacterial effect of the phage cocktail or phage cocktail-sub-minimal inhibitory concentration (MIC) of kasugamycin was significantly higher than the kasugamycin at the MIC. The selected phages were experimentally stable under environmental factors such as thermal or pH stress. Genomic analysis revealed these are novel Erwinia-infecting phages, and did not encode antibiotic-, virulence-, or lysogenic phage-related genes. In conclusion, we suggest the potential of the phage cocktail and kasugamycin combination as an effective strategy that would minimize the use of antibiotics, which are being excessively used in order to control fire blight pathogens.
Keywords: Erwinia amylovora; bacteriophage; fire blight; phage therapy; phage–antibiotic synergy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures










References
Grants and funding
LinkOut - more resources
Full Text Sources

