Roles of Ferredoxin-NADP+ Oxidoreductase and Flavodoxin in NAD(P)H-Dependent Electron Transfer Systems
- PMID: 36358515
- PMCID: PMC9687028
- DOI: 10.3390/antiox11112143
Roles of Ferredoxin-NADP+ Oxidoreductase and Flavodoxin in NAD(P)H-Dependent Electron Transfer Systems
Abstract
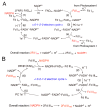
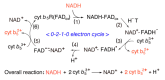
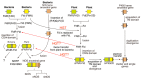
Distinct isoforms of FAD-containing ferredoxin-NADP+ oxidoreductase (FNR) and ferredoxin (Fd) are involved in photosynthetic and non-photosynthetic electron transfer systems. The FNR (FAD)-Fd [2Fe-2S] redox pair complex switches between one- and two-electron transfer reactions in steps involving FAD semiquinone intermediates. In cyanobacteria and some algae, one-electron carrier Fd serves as a substitute for low-potential FMN-containing flavodoxin (Fld) during growth under low-iron conditions. This complex evolves into the covalent FNR (FAD)-Fld (FMN) pair, which participates in a wide variety of NAD(P)H-dependent metabolic pathways as an electron donor, including bacterial sulfite reductase, cytochrome P450 BM3, plant or mammalian cytochrome P450 reductase and nitric oxide synthase isoforms. These electron transfer systems share the conserved Ser-Glu/Asp pair in the active site of the FAD module. In addition to physiological electron acceptors, the NAD(P)H-dependent diflavin reductase family catalyzes a one-electron reduction of artificial electron acceptors such as quinone-containing anticancer drugs. Conversely, NAD(P)H: quinone oxidoreductase (NQO1), which shares a Fld-like active site, functions as a typical two-electron transfer antioxidant enzyme, and the NQO1 and UDP-glucuronosyltransfease/sulfotransferase pairs function as an antioxidant detoxification system. In this review, the roles of the plant FNR-Fd and FNR-Fld complex pairs were compared to those of the diflavin reductase (FAD-FMN) family. In the final section, evolutionary aspects of NAD(P)H-dependent multi-domain electron transfer systems are discussed.
Keywords: catalytic cycle; diflavin reductase family; electron transfer; evolutionary aspects; ferredoxin (Fd); ferredoxin-NADP+ oxidoreductase (FNR); flavodoxin (Fld); redox potentials.
Conflict of interest statement
The author has no conflict of interest to declare.
Figures











Similar articles
-
A theoretical multiscale treatment of protein-protein electron transfer: The ferredoxin/ferredoxin-NADP(+) reductase and flavodoxin/ferredoxin-NADP(+) reductase systems.Biochim Biophys Acta. 2015 Dec;1847(12):1530-8. doi: 10.1016/j.bbabio.2015.09.002. Epub 2015 Sep 16. Biochim Biophys Acta. 2015. PMID: 26385068
-
Effects of chemical modification of Anabaena flavodoxin and ferredoxin-NADP+ reductase on the kinetics of interprotein electron transfer reactions.Eur J Biochem. 1992 Dec 1;210(2):577-83. doi: 10.1111/j.1432-1033.1992.tb17457.x. Eur J Biochem. 1992. PMID: 1459139
-
Interaction of Ferredoxin-NADP(+) Reductase with its Substrates: Optimal Interaction for Efficient Electron Transfer.Photosynth Res. 2004 Feb;79(2):113-31. doi: 10.1023/B:PRES.0000015386.67746.2c. Photosynth Res. 2004. PMID: 16228387
-
Evolution of the acceptor side of photosystem I: ferredoxin, flavodoxin, and ferredoxin-NADP+ oxidoreductase.Photosynth Res. 2017 Dec;134(3):235-250. doi: 10.1007/s11120-017-0338-2. Epub 2017 Feb 1. Photosynth Res. 2017. PMID: 28150152 Review.
-
Flavin-Based Electron Bifurcation, Ferredoxin, Flavodoxin, and Anaerobic Respiration With Protons (Ech) or NAD+ (Rnf) as Electron Acceptors: A Historical Review.Front Microbiol. 2018 Mar 14;9:401. doi: 10.3389/fmicb.2018.00401. eCollection 2018. Front Microbiol. 2018. PMID: 29593673 Free PMC article. Review.
Cited by
-
Metabolic Engineering of Model Microorganisms for the Production of Xanthophyll.Microorganisms. 2023 May 9;11(5):1252. doi: 10.3390/microorganisms11051252. Microorganisms. 2023. PMID: 37317226 Free PMC article. Review.
-
The iron chelator pulcherriminic acid mediates the light response in Bacillus subtilis biofilms.Nat Commun. 2025 Jul 1;16(1):5446. doi: 10.1038/s41467-025-60560-4. Nat Commun. 2025. PMID: 40595503 Free PMC article.
-
Redox Properties of Bacillus subtilis Ferredoxin:NADP+ Oxidoreductase: Potentiometric Characteristics and Reactions with Pro-Oxidant Xenobiotics.Int J Mol Sci. 2024 May 14;25(10):5373. doi: 10.3390/ijms25105373. Int J Mol Sci. 2024. PMID: 38791410 Free PMC article.
-
Transcriptomic and proteomic analyses of Mangifera indica in response to Xanthomonas critis pv. mangiferaeindicae.Front Microbiol. 2023 Jul 4;14:1220101. doi: 10.3389/fmicb.2023.1220101. eCollection 2023. Front Microbiol. 2023. PMID: 37469435 Free PMC article.
-
Transcriptomic and proteomic-based analysis of the mechanisms by which drought and salt stresses affect the quality of Isatidis Folium.BMC Plant Biol. 2025 Mar 14;25(1):332. doi: 10.1186/s12870-025-06309-z. BMC Plant Biol. 2025. PMID: 40087613 Free PMC article.
References
-
- Shinohara F., Kurisu G., Hanke G., Bowsher C., Hase T., Kimata-Ariga Y. Structural basis for the isotype-specific interactions of ferredoxin and ferredoxin: NADP+ oxidoreductase: An evolutionary switch between photosynthetic and heterotrophic assimilation. Photosynth. Res. 2017;134:281–289. doi: 10.1007/s11120-016-0331-1. - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

