Inhibition of IκB Kinase Is a Potential Therapeutic Strategy to Circumvent Resistance to Epidermal Growth Factor Receptor Inhibition in Triple-Negative Breast Cancer Cells
- PMID: 36358633
- PMCID: PMC9654813
- DOI: 10.3390/cancers14215215
Inhibition of IκB Kinase Is a Potential Therapeutic Strategy to Circumvent Resistance to Epidermal Growth Factor Receptor Inhibition in Triple-Negative Breast Cancer Cells
Abstract
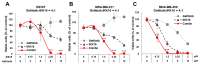
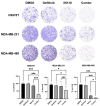
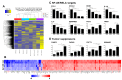
Triple-negative breast cancer (TNBC) remains as an intractable malignancy with limited therapeutic targets. High expression of epidermal growth factor receptor (EGFR) has been associated with a poor prognosis of TNBC; however, EGFR targeting has failed with unfavorable clinical outcomes. Here, we performed a combinatorial screening of fifty-five protein kinase inhibitors with the EGFR inhibitor gefitinib in the TNBC cell line MDA-MB-231 and identified the IκB kinase (IKK) inhibitor IKK16 as a sensitizer of gefitinib. Cell viability and clonogenic survival assays were performed to evaluate the antiproliferative effects of the gefitinib and IKK16 (Gefitinib + IKK16) combination in TNBC cell lines. Western blot analyses were also performed to reveal the potential mode of action of this combination. In addition, next-generation sequencing (NGS) analysis was performed in Gefitinib+IKK16-treated cells. The Gefitinib+IKK16 treatment synergistically reduced cell viability and colony formation of TNBC cell lines such as HS578T, MDA-MB-231, and MDA-MB-468. This combination downregulated p-STAT3, p-AKT, p-mTOR, p-GSK3β, and p-RPS6. In addition, p-NF-κB and the total NF-κB were also regulated by this combination. Furthermore, NGS analysis revealed that NF-κB/RELA targets including CCL2, CXCL8, EDN1, IL-1β, IL-6, and SERPINE1 were further reduced and several potential tumor suppressors, such as FABP3, FADS2, FDFT1, SEMA6A, and PCK2, were synergistically induced by the Gefitinib-+IKK16 treatment. Taken together, we identified the IKK/NF-κB pathway as a potential target in combination of EGFR inhibition for treating TNBC.
Keywords: IκB kinase (IKK) inhibition; anticancer; combination; epidermal growth factor receptor (EGFR) inhibition; gefitinib; kinase inhibitor; resistance; synergism; triple-negative breast cancer (TNBC).
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Grants and funding
- NRF-2022R1l1A1A01064022/National Research Foundation of Korea
- NRF-2019R1l1A3A01059102/National Research Foundation of Korea
- NRF-2021R1A5A2022318/National Research Foundation of Korea
- NRF-2021R1I1A2054560/National Research Foundation of Korea
- NRF-2021R1A6A3A01086886/National Research Foundation of Korea
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

