Hepcidin Upregulation in Colorectal Cancer Associates with Accumulation of Regulatory Macrophages and Epithelial-Mesenchymal Transition and Correlates with Progression of the Disease
- PMID: 36358713
- PMCID: PMC9658525
- DOI: 10.3390/cancers14215294
Hepcidin Upregulation in Colorectal Cancer Associates with Accumulation of Regulatory Macrophages and Epithelial-Mesenchymal Transition and Correlates with Progression of the Disease
Abstract
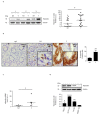
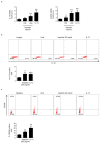
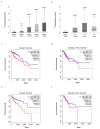
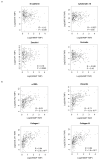
Advanced, metastatic colorectal cancer (CRC) is associated with high rate of mortality because of its poor responsiveness to chemotherapy/immunotherapy. Recent studies have shown that hepcidin, a peptide hormone produced mainly by hepatocytes, is expressed by and enhances the growth of tumor cells. We here assessed whether hepcidin expression helps identify subsets of CRC with advanced and aggressive course. By integrating results of in vitro/ex vivo studies with data of bioinformatics databases, we initially showed that hepcidin RNA and protein expression was more pronounced in tissue samples taken from the tumor area, as compared to the macroscopically unaffected, adjacent, colonic mucosa of CRC patients. The induction of hepcidin in the colonic epithelial cell line HCEC-1ct by interleukin (IL)-6, IL-21 and IL-23 occurred via a Stat3-dependent mechanism and, in primary CRC cells, hepcidin co-localized with active Stat3. In CRC tissue, hepcidin content correlated mainly with macrophage accumulation and IL-10 and CD206 expression, two markers of regulatory macrophages. Consistently, both IL-10 and CD206 were up-regulated by hepcidin in blood mononuclear cells. The highest levels of hepcidin were found in metastatic CRC and survival analysis showed that high expression of hepcidin associated with poor prognosis. Moreover, hepcidin expression correlated with markers of epithelial-to-mesenchymal transition and the silencing of hepcidin in CRC cells reduced epithelial-to-mesenchymal transition markers. These findings indicate that hepcidin is markedly induced in the advanced stages of CRC and suggest that it could serve as a prognostic biomarker in CRC.
Keywords: Stat3; cancer metastasis; cytokines; prognosis; type 2 macrophages.
Conflict of interest statement
G.M. served as a consultant for First Wave BioPharma and filed a patent related to the treatment of inflammatory bowel diseases with Smad7 antisense oligonucleotides. The other authors have no conflict of interest.
Figures




Similar articles
-
Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis.Mol Cancer. 2019 Mar 30;18(1):64. doi: 10.1186/s12943-019-0976-4. Mol Cancer. 2019. PMID: 30927925 Free PMC article.
-
CPEB3 inhibits epithelial-mesenchymal transition by disrupting the crosstalk between colorectal cancer cells and tumor-associated macrophages via IL-6R/STAT3 signaling.J Exp Clin Cancer Res. 2020 Jul 11;39(1):132. doi: 10.1186/s13046-020-01637-4. J Exp Clin Cancer Res. 2020. PMID: 32653013 Free PMC article.
-
Hepcidin Is a Valuable Therapeutic Target for Colorectal Cancer.Cancers (Basel). 2024 Dec 5;16(23):4068. doi: 10.3390/cancers16234068. Cancers (Basel). 2024. PMID: 39682254 Free PMC article. Review.
-
Aberrantly expressed Fra-1 by IL-6/STAT3 transactivation promotes colorectal cancer aggressiveness through epithelial-mesenchymal transition.Carcinogenesis. 2015 Apr;36(4):459-68. doi: 10.1093/carcin/bgv017. Epub 2015 Mar 6. Carcinogenesis. 2015. PMID: 25750173 Free PMC article.
-
Inhibitor of growth 4 suppresses colorectal cancer growth and invasion by inducing G1 arrest, inhibiting tumor angiogenesis and reversing epithelial-mesenchymal transition.Oncol Rep. 2016 May;35(5):2927-35. doi: 10.3892/or.2016.4626. Epub 2016 Feb 18. Oncol Rep. 2016. PMID: 26936485
Cited by
-
Managing the Dual Nature of Iron to Preserve Health.Int J Mol Sci. 2023 Feb 16;24(4):3995. doi: 10.3390/ijms24043995. Int J Mol Sci. 2023. PMID: 36835406 Free PMC article. Review.
-
Soluble Hemojuvelin and Ferritin: Potential Prognostic Markers in Pediatric Hematopoietic Cell Transplantation.Cancers (Basel). 2023 Feb 7;15(4):1041. doi: 10.3390/cancers15041041. Cancers (Basel). 2023. PMID: 36831385 Free PMC article.
-
Targeting hepcidin in colorectal cancer triggers a TNF-dependent-gasdermin E-driven immunogenic cell death response.Exp Hematol Oncol. 2024 Sep 27;13(1):95. doi: 10.1186/s40164-024-00562-y. Exp Hematol Oncol. 2024. PMID: 39334507 Free PMC article.
-
Enhanced CRC Growth in Iron-Rich Environment, Facts and Speculations.Int J Mol Sci. 2024 Nov 19;25(22):12389. doi: 10.3390/ijms252212389. Int J Mol Sci. 2024. PMID: 39596454 Free PMC article. Review.
-
Multifaceted Insights into Innovative Approaches to Treating Colorectal Cancer Metastasis: From Emerging Biological Factors to Radiomics.Cancers (Basel). 2023 Sep 20;15(18):4644. doi: 10.3390/cancers15184644. Cancers (Basel). 2023. PMID: 37760613 Free PMC article.
References
-
- Fedirko V., Tramacere I., Bagnardi V., Rota M., Scotti L., Islami F., Negri E., Straif K., Romieu I., La Vecchia C., et al. Alcohol drinking and colorectal cancer risk: An overall and dose-response meta-analysis of published studies. Ann. Oncol. 2011;22:1958–1972. doi: 10.1093/annonc/mdq653. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

