Improving Precise Genome Editing Using Donor DNA/gRNA Hybrid Duplex Generated by Complementary Bases
- PMID: 36358971
- PMCID: PMC9687273
- DOI: 10.3390/biom12111621
Improving Precise Genome Editing Using Donor DNA/gRNA Hybrid Duplex Generated by Complementary Bases
Abstract
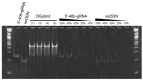
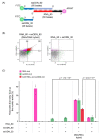
In precise genome editing, site-specific DNA double-strand breaks (DSBs) induced by the CRISPR/Cas9 system are repaired via homology-directed repair (HDR) using exogenous donor DNA templates. However, the low efficiency of HDR-mediated genome editing is a barrier to widespread use. In this study, we created a donor DNA/guide RNA (gRNA) hybrid duplex (DGybrid) that was composed of sequence-extended gRNA and single-stranded oligodeoxynucleotide (ssODN) combined with complementary bases without chemical modifications to increase the concentration of donor DNA at the cleavage site. The efficiency of genome editing using DGybrid was evaluated in Saccharomyces cerevisiae. The results show a 1.8-fold (from 35% to 62%) improvement in HDR-mediated editing efficiency compared to genome editing in which gRNA and donor DNA were introduced separately. In addition, analysis of the nucleic acid introduction efficiency using flow cytometry indicated that both RNA and ssODNs are efficiently incorporated into cells together by using the DNA/RNA hybrid. Our technique would be preferred as a universal and concise tool for improving the efficiency of HDR-mediated genome editing.
Keywords: CRISPR/Cas9; DNA/RNA hybrid; Saccharomyces cerevisiae; genome editing; guide RNA; single-stranded oligodeoxynucleotide.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the study design; collection, analyses, or interpretation of data; writing of the manuscript; or decision to publish the results.
Figures



References
-
- DeWitt M.A., Magis W., Bray N.L., Wang T., Berman J.R., Urbinati F., Heo S.J., Mitros T., Munoz D.P., Boffelli D., et al. Selection-free Genome Editing of the Sickle Mutation in Human Adult Hematopoietic Stem/progenitor Cells. Sci. Transl. Med. 2016;8:360ra134. doi: 10.1126/scitranslmed.aaf9336. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

