Transcriptome Sequencing Reveals Pathways Related to Proliferation and Differentiation of Shitou Goose Myoblasts
- PMID: 36359079
- PMCID: PMC9658593
- DOI: 10.3390/ani12212956
Transcriptome Sequencing Reveals Pathways Related to Proliferation and Differentiation of Shitou Goose Myoblasts
Abstract
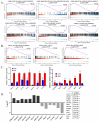
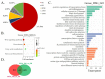
Chinese Shitou goose is a type of large goose with high meat yield. Understanding the genetic regulation of muscle development in Shitou goose would be beneficial to improve the meat production traits of geese. Muscle development is regulated by genes related to myoblast proliferation and differentiation. In this study, the RNA-seq method was used to construct the mRNA and lncRNA expression profiles of Shitou goose myoblasts and myotubes. A total of 1664 differentially expressed (DE) mRNAs and 244 DE-lncRNAs were identified. The alternative mRNA splicing in proliferation and differentiation stages was also analyzed. Notably, pathways enriched in DE-mRNAs, DE-splicing transcripts, and DE-lncRNAs all point to the Wnt signaling pathway, indicating that the Wnt signaling is a key regulatory pathway of muscle development in Shitou goose. We also constructed the interactive network of DE-lncRNAs and DE-mRNAs and revealed some key genes of lncRNAs regulating the proliferation and differentiation of myoblasts. These results provide new insights for the study of the muscle development of the Shitou goose.
Keywords: RNA-seq; Shitou goose; alternative splicing; long non-coding RNAs; mRNA.
Conflict of interest statement
All authors declare no conflict of interest.
Figures




References
-
- Ye C., Zhong R. Study on the growth model of lion headed geese. J. Shihezi Univ. (Nat. Sci. Ed.) 2005;7:694–697.
Grants and funding
- Grant No. 2020B020222003/Key-Area Research and Development Program of Guangdong Province
- Grant No. 31972544/Natural Scientific Foundation of China
- Grant No. 2019TQ05N470/Guangdong Special Branch Plans of Young Talent with Scientific and Technological Innovation
- Grant No. 2019A1515010923/Natural Science Foundation of Guangdong Province, China
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

