Adenovirus-Inspired Virus-Like-Particles Displaying Melanoma Tumor Antigen Specifically Target Human DC Subsets and Trigger Antigen-Specific Immune Responses
- PMID: 36359404
- PMCID: PMC9687312
- DOI: 10.3390/biomedicines10112881
Adenovirus-Inspired Virus-Like-Particles Displaying Melanoma Tumor Antigen Specifically Target Human DC Subsets and Trigger Antigen-Specific Immune Responses
Abstract
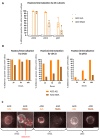
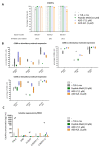
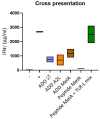
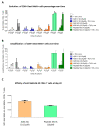
Virus-like particles constitute versatile vectors that can be used as vaccine platforms in many fields from infectiology and more recently to oncology. We previously designed non-infectious adenovirus-inspired 60-mer dodecahedric virus-like particles named ADDomers displaying on their surface either a short epitope or a large tumor/viral antigen. In this work, we explored for the first time the immunogenicity of ADDomers exhibiting melanoma-derived tumor antigen/epitope and their impact on the features of human dendritic cell (DC) subsets. We first demonstrated that ADDomers displaying tumor epitope/antigen elicit a strong immune-stimulating potential of human DC subsets (cDC2s, cDC1s, pDCs), which were able to internalize and cross-present tumor antigen, and subsequently cross-prime antigen-specific T-cell responses. To further limit off-target effects and enhance DC targeting, we engineered specific motifs to de-target epithelial cells and improve DCs' addressing. The improved engineered platform making it possible to display large antigen represents a tool to overcome the barrier of immune allele restriction, broadening the immune response, and paving the way to its potential utilization in humans as an off-the-shelf vaccine.
Keywords: C-type lectin receptors; adenovirus; immunotherapy; melanoma; vaccine platform.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

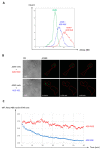
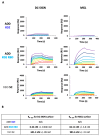


References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

