A Simplified Sanger Sequencing Method for Detection of Relevant SARS-CoV-2 Variants
- PMID: 36359452
- PMCID: PMC9689870
- DOI: 10.3390/diagnostics12112609
A Simplified Sanger Sequencing Method for Detection of Relevant SARS-CoV-2 Variants
Abstract
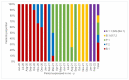
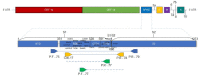
Molecular surveillance of the new coronavirus through new genomic sequencing technologies revealed the circulation of important variants of SARS-CoV-2. Sanger sequencing has been useful in identifying important variants of SARS-CoV-2 without the need for whole-genome sequencing. A sequencing protocol was constructed to cover a region of 1000 base pairs, from a 1120 bp product generated after a two-step RT-PCR assay in samples positive for SARS-CoV-2. Consensus sequence construction and mutation identification were performed. Of all 103 samples sequenced, 69 contained relevant variants represented by 20 BA.1, 13 delta, 22 gamma, and 14 zeta, identified between June 2020 and February 2022. All sequences found were aligned with representative sequences of the variants. Using the Sanger sequencing methodology, we were able to develop a more accessible protocol to assist viral surveillance with a more accessible platform.
Keywords: SARS-CoV-2; Sanger; surveillance; variants.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Chand M., Hopkins S., Dabrera G., Achison C., Barclay W., Ferguson N., Volz E., Loman N., Rambaut A., Barrett J. Investigation of novel SARS-COV-2 variant Variant of Concern 202012/01. [(accessed on 10 May 2022)]; Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploa....
LinkOut - more resources
Full Text Sources
Miscellaneous

