Whole Transcriptome Sequencing Reveals Cancer-Related, Prognostically Significant Transcripts and Tumor-Infiltrating Immunocytes in Mantle Cell Lymphoma
- PMID: 36359790
- PMCID: PMC9654955
- DOI: 10.3390/cells11213394
Whole Transcriptome Sequencing Reveals Cancer-Related, Prognostically Significant Transcripts and Tumor-Infiltrating Immunocytes in Mantle Cell Lymphoma
Abstract
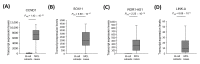
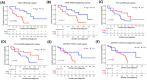
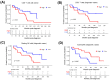
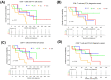
Mantle cell lymphoma (MCL) is an aggressive B-cell non-Hodgkin lymphoma (NHL) subtype characterized by overexpression of CCND1 and SOX11 genes. It is generally associated with clinically poor outcomes despite recent improvements in therapeutic approaches. The genes associated with the development and prognosis of MCL are still largely unknown. Through whole transcriptome sequencing (WTS), we identified mRNAs, lncRNAs, and alternative transcripts differentially expressed in MCL cases compared with reactive tonsil B-cell subsets. CCND1, VCAM1, and VWF mRNAs, as well as MIR100HG and ROR1-AS1 lncRNAs, were among the top 10 most significantly overexpressed, oncogenesis-related transcripts. Survival analyses with each of the top upregulated transcripts showed that MCL cases with high expression of VWF mRNA and low expression of FTX lncRNA were associated with poor overall survival. Similarly, high expression of MSTRG.153013.3, an overexpressed alternative transcript, was associated with shortened MCL survival. Known tumor suppressor candidates (e.g., PI3KIP1, UBXN) were significantly downregulated in MCL cases. Top differentially expressed protein-coding genes were enriched in signaling pathways related to invasion and metastasis. Survival analyses based on the abundance of tumor-infiltrating immunocytes estimated with CIBERSORTx showed that high ratios of CD8+ T-cells or resting NK cells and low ratios of eosinophils are associated with poor overall survival in diagnostic MCL cases. Integrative analysis of tumor-infiltrating CD8+ T-cell abundance and overexpressed oncogene candidates showed that MCL cases with high ratio CD8+ T-cells and low expression of FTX or PCA3 can potentially predict high-risk MCL patients. WTS results were cross-validated with qRT-PCR of selected transcripts as well as linear correlation analyses. In conclusion, expression levels of oncogenesis-associated transcripts and/or the ratios of microenvironmental immunocytes in MCL tumors may be used to improve prognostication, thereby leading to better patient management and outcomes.
Keywords: WTS; alternative transcript; lncRNA; mRNA; mantle cell lymphoma; oncogene; prognosis; tumor microenvironment; tumor suppressor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Veloza L., Ribera-Cortada I., Campo E. Mantle Cell Lymphoma Biology. Clin. Lymphoma Myeloma Leuk. 2018;18:S97–S98. doi: 10.1016/j.clml.2018.06.070. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

