Identification of the Telomere elongation Mutation in Drosophila
- PMID: 36359878
- PMCID: PMC9659042
- DOI: 10.3390/cells11213484
Identification of the Telomere elongation Mutation in Drosophila
Abstract
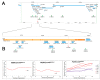
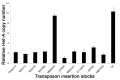
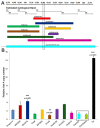
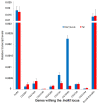
Telomeres in Drosophila melanogaster, which have inspired a large part of Sergio Pimpinelli work, are similar to those of other eukaryotes in terms of their function. Yet, their length maintenance relies on the transposition of the specialized retrotransposons Het-A, TART, and TAHRE, rather than on the activity of the enzyme telomerase as it occurs in most other eukaryotic organisms. The length of the telomeres in Drosophila thus depends on the number of copies of these transposable elements. Our previous work has led to the isolation of a dominant mutation, Tel1, that caused a several-fold elongation of telomeres. In this study, we molecularly identified the Tel1 mutation by a combination of transposon-induced, site-specific recombination and next-generation sequencing. Recombination located Tel1 to a 15 kb region in 92A. Comparison of the DNA sequence in this region with the Drosophila Genetic Reference Panel of wild-type genomic sequences delimited Tel1 to a 3 bp deletion inside intron 8 of Ino80. Furthermore, CRISPR/Cas9-induced deletions surrounding the same region exhibited the Tel1 telomere phenotype, confirming a strict requirement of this intron 8 gene sequence for a proper regulation of Drosophila telomere length.
Keywords: Drosophila melanogaster; next-generation sequencing; telomere; transposon-induced recombination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Frydrychova R.C., Mason J.M. Telomeres: Their Structure and Maintenance. In: Stuart D., editor. The Mechaisms of DNA Replication. InTech, Open Access Publisher; Rijeka, Croatia: 2013. pp. 423–443.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

