Bioinformatics Analysis and Validation of the Role of Lnc-RAB11B-AS1 in the Development and Prognosis of Hepatocellular Carcinoma
- PMID: 36359911
- PMCID: PMC9657516
- DOI: 10.3390/cells11213517
Bioinformatics Analysis and Validation of the Role of Lnc-RAB11B-AS1 in the Development and Prognosis of Hepatocellular Carcinoma
Abstract
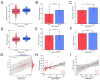
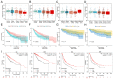
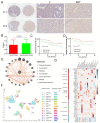
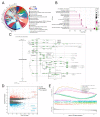
Lnc-RAB11B-AS1 is reported to be dysregulated in several types of cancers and can function as both an oncogene and tumor suppressor gene. To evaluate the potential role of lnc-RAB11B-AS1 in hepatocellular carcinoma (HCC), we investigated and evaluated its expression in HCC based on the data mining of a series of public databases, including TCGA, GEO, ICGC, HPA, DAVID, cBioPortal, GeneMIANA, TIMER, and ENCORI. The data showed downregulation of lnc-RAB11B-AS1 in HCC and was accompanied by the synchronous downregulation of the targeted RAB11B mRNA and its protein. Low expression of lnc-RAB11B-AS1 was associated with shorter overall survival (OS) and disease-free survival (DFS) of HCC patients, PD1/PD-L1 was correlated with low expression of RAB11B. Furthermore, Gene Ontology (GO) functional annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis showed a correlation between immune cell change and non-alcoholic fatty liver disease. The above findings revealed that lnc-RAB11B-AS1 was down-regulated in HCC and closely associated with the clinical stage of the HCC patients, suggesting that lnc-RAB11B-AS1 could be a possible predictor for HCC and a potential new therapeutic target for the treatment of HCC.
Keywords: RAB11B; hepatocellular carcinoma (HCC); lnc-RAB11B-AS1; prognosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
METTL16 promotes hepatocellular carcinoma progression through downregulating RAB11B-AS1 in an m6A-dependent manner.Cell Mol Biol Lett. 2022 May 20;27(1):41. doi: 10.1186/s11658-022-00342-8. Cell Mol Biol Lett. 2022. PMID: 35596159 Free PMC article.
-
RNA sequencing reveals the long noncoding RNA and mRNA profiles and identifies long non-coding RNA TSPAN12 as a potential microvascular invasion-related biomarker in hepatocellular carcinoma.Biomed Pharmacother. 2020 Jun;126:110111. doi: 10.1016/j.biopha.2020.110111. Epub 2020 Mar 26. Biomed Pharmacother. 2020. PMID: 32222644
-
LincRNA ZNF529-AS1 inhibits hepatocellular carcinoma via FBXO31 and predicts the prognosis of hepatocellular carcinoma patients.BMC Bioinformatics. 2023 Feb 17;24(1):54. doi: 10.1186/s12859-023-05189-0. BMC Bioinformatics. 2023. PMID: 36803542 Free PMC article.
-
Upregulation of long noncoding RNA RAB11B-AS1 promotes tumor metastasis and predicts poor prognosis in lung cancer.Ann Transl Med. 2020 May;8(9):582. doi: 10.21037/atm.2020.04.52. Ann Transl Med. 2020. PMID: 32566609 Free PMC article.
-
TP73-AS1 as a predictor of clinicopathological parameters and prognosis in human malignancies: a meta and bioinformatics analysis.BMC Cancer. 2022 May 25;22(1):581. doi: 10.1186/s12885-022-09658-2. BMC Cancer. 2022. PMID: 35614413 Free PMC article.
Cited by
-
The current status and future of PD-L1 in liver cancer.Front Immunol. 2023 Dec 12;14:1323581. doi: 10.3389/fimmu.2023.1323581. eCollection 2023. Front Immunol. 2023. PMID: 38155974 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

