The Complex Dynamic of Phase I Drug Metabolism in the Early Stages of Doxorubicin Resistance in Breast Cancer Cells
- PMID: 36360213
- PMCID: PMC9689592
- DOI: 10.3390/genes13111977
The Complex Dynamic of Phase I Drug Metabolism in the Early Stages of Doxorubicin Resistance in Breast Cancer Cells
Abstract
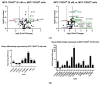
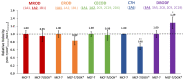
The altered activity of drug metabolism enzymes (DMEs) is a hallmark of chemotherapy resistance. Cytochrome P450s (CYPs), mainly CYP3A4, and several oxidoreductases are responsible for Phase I metabolism of doxorubicin (DOX), an anthracycline widely used in breast cancer (BC) treatment. This study aimed to investigate the role of Phase I DMEs involved in the first stages of acquisition of DOX-resistance in BC cells. For this purpose, the expression of 92 DME genes and specific CYP-complex enzymes activities were assessed in either sensitive (MCF-7 parental cells; MCF-7/DOXS) or DOX-resistant (MCF-7/DOXR) cells. The DMEs genes detected to be significantly differentially expressed in MCF-7/DOXR cells (12 CYPs and eight oxidoreductases) were indicated previously to be involved in tumor progression and/or chemotherapy response. The analysis of CYP-mediated activities suggests a putative enhanced CYP3A4-dependent metabolism in MCF-7/DOXR cells. A discrepancy was observed between CYP-enzyme activities and their corresponding levels of mRNA transcripts. This is indicative that the phenotype of DMEs is not linearly correlated with transcription induction responses, confirming the multifactorial complexity of this mechanism. Our results pinpoint the potential role of specific CYPs and oxidoreductases involved in the metabolism of drugs, retinoic and arachidonic acids, in the mechanisms of chemo-resistance to DOX and carcinogenesis of BC.
Keywords: breast cancer (BC); cytochrome P450 (CYP); doxorubicin (DOX); drug metabolism enzymes (DMEs); drug resistance (DR); oxidoreductases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Li Y., Steppi A., Zhou Y., Mao F., Miller P.C., He M.M., Zhao T., Sun Q., Zhang J. Tumoral Expression of Drug and Xenobiotic Metabolizing Enzymes in Breast Cancer Patients of Different Ethnicities with Implications to Personalized Medicine. Sci. Rep. 2017;7:4747. doi: 10.1038/s41598-017-04250-2. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

