CircRNAs Related to Breast Muscle Development and Their Interaction Regulatory Network in Gushi Chicken
- PMID: 36360215
- PMCID: PMC9689937
- DOI: 10.3390/genes13111974
CircRNAs Related to Breast Muscle Development and Their Interaction Regulatory Network in Gushi Chicken
Abstract
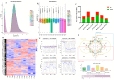
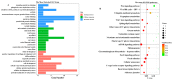
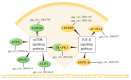
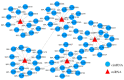
Circular RNAs (circRNAs) play a significant regulatory role during skeletal muscle development. To identify circRNAs during postnatal skeletal muscle development in chickens, we constructed 12 cDNA libraries from breast muscle tissues of Chinese Gushi chickens at 6, 14, 22, and 30 weeks and performed RNA sequencing. In total, 2112 circRNAs were identified, and among them 79.92% were derived from exons. CircRNAs are distributed on all chromosomes of chickens, especially chromosomes 1-9 and Z. Bioinformatics analysis showed that each circRNA had an average of 38 miRNA binding sites, 61.32% of which have internal ribosomal entry site (IRES) elements. Furthermore, in total 543 differentially expressed circRNAs (DE-circRNAs) were identified. Functional enrichment analysis revealed that DE-circRNAs source genes are engaged in biological processes and muscle development-related pathways; for example, cell differentiation, sarcomere, and myofibril formation, mTOR signaling pathway, and TGF-β signaling pathway, etc. We also established a competitive endogenous RNA (ceRNA) regulatory network associated with skeletal muscle development. The results in this report indicate that circRNAs can mediate the development of chicken skeletal muscle by means of a complex ceRNA network among circRNAs, miRNAs, genes, and pathways. The findings of this study might help increase the number of known circRNAs in skeletal muscle tissue and offer a worthwhile resource to further investigate the function of circRNAs in chicken skeletal muscle development.
Keywords: Gushi chickens; ceRNA; circular RNAs; regulatory network; skeletal muscle.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
Characteristics and expression profiles of circRNAs during abdominal adipose tissue development in Chinese Gushi chickens.PLoS One. 2021 Apr 15;16(4):e0249288. doi: 10.1371/journal.pone.0249288. eCollection 2021. PLoS One. 2021. PMID: 33857153 Free PMC article.
-
Integrative analysis of circRNA, miRNA, and mRNA profiles to reveal ceRNA regulation in chicken muscle development from the embryonic to post-hatching periods.BMC Genomics. 2022 May 3;23(1):342. doi: 10.1186/s12864-022-08525-5. BMC Genomics. 2022. PMID: 35505302 Free PMC article.
-
Genome-wide analysis of circular RNA-mediated ceRNA regulation in porcine skeletal muscle development.BMC Genomics. 2023 Apr 12;24(1):196. doi: 10.1186/s12864-023-09284-7. BMC Genomics. 2023. PMID: 37046223 Free PMC article.
-
Circular RNAs and host genes act synergistically in regulating cellular processes and functions in skeletal myogenesis.Gene. 2025 Mar 10;940:149189. doi: 10.1016/j.gene.2024.149189. Epub 2024 Dec 24. Gene. 2025. PMID: 39724991 Review.
-
Regulation of Non-Coding RNA in the Growth and Development of Skeletal Muscle in Domestic Chickens.Genes (Basel). 2022 Jun 9;13(6):1033. doi: 10.3390/genes13061033. Genes (Basel). 2022. PMID: 35741795 Free PMC article. Review.
Cited by
-
RNA-Seq Analysis Revealed circRNAs and Genes Associated with Abdominal Fat Deposition in Ducks.Animals (Basel). 2024 Jan 14;14(2):260. doi: 10.3390/ani14020260. Animals (Basel). 2024. PMID: 38254429 Free PMC article.
-
Multi-Omics Profiling of Lipid Variation and Regulatory Mechanisms in Poultry Breast Muscles.Animals (Basel). 2025 Feb 27;15(5):694. doi: 10.3390/ani15050694. Animals (Basel). 2025. PMID: 40075981 Free PMC article.
-
miR-200b-3p affects the proliferation and differentiation of chicken preadipocytes by modulating SESN1 expression through competition with CircADGRF5.Poult Sci. 2025 May;104(5):105068. doi: 10.1016/j.psj.2025.105068. Epub 2025 Mar 19. Poult Sci. 2025. PMID: 40132317 Free PMC article.
-
Pharmacokinetics of Danofloxacin in Gushi Chickens after Single Oral and Intravenous Administration.Metabolites. 2023 Aug 2;13(8):906. doi: 10.3390/metabo13080906. Metabolites. 2023. PMID: 37623849 Free PMC article.
-
Emerging roles of circular RNAs on the regulation of production traits in chicken.Poult Sci. 2025 Jan;104(1):104612. doi: 10.1016/j.psj.2024.104612. Epub 2024 Nov 28. Poult Sci. 2025. PMID: 39647355 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

