Bi-EB: Empirical Bayesian Biclustering for Multi-Omics Data Integration Pattern Identification among Species
- PMID: 36360219
- PMCID: PMC9690013
- DOI: 10.3390/genes13111982
Bi-EB: Empirical Bayesian Biclustering for Multi-Omics Data Integration Pattern Identification among Species
Abstract
Although several biclustering algorithms have been studied, few are used for cross-pattern identification across species using multi-omics data mining. A fast empirical Bayesian biclustering (Bi-EB) algorithm is developed to detect the patterns shared from both integrated omics data and between species. The Bi-EB algorithm addresses the clinical critical translational question using the bioinformatics strategy, which addresses how modules of genotype variation associated with phenotype from cancer cell screening data can be identified and how these findings can be directly translated to a cancer patient subpopulation. Empirical Bayesian probabilistic interpretation and ratio strategy are proposed in Bi-EB for the first time to detect the pairwise regulation patterns among species and variations in multiple omics on a gene level, such as proteins and mRNA. An expectation-maximization (EM) optimal algorithm is used to extract the foreground co-current variations out of its background noise data by adjusting parameters with bicluster membership probability threshold Ac; and the bicluster average probability p. Three simulation experiments and two real biology mRNA and protein data analyses conducted on the well-known Cancer Genomics Atlas (TCGA) and The Cancer Cell Line Encyclopedia (CCLE) verify that the proposed Bi-EB algorithm can significantly improve the clustering recovery and relevance accuracy, outperforming the other seven biclustering methods-Cheng and Church (CC), xMOTIFs, BiMax, Plaid, Spectral, FABIA, and QUBIC-with a recovery score of 0.98 and a relevance score of 0.99. At the same time, the Bi-EB algorithm is used to determine shared the causality patterns of mRNA to the protein between patients and cancer cells in TCGA and CCLE breast cancer. The clinically well-known treatment target protein module estrogen receptor (ER), ER (p118), AR, BCL2, cyclin E1, and IGFBP2 are identified in accordance with their mRNA expression variations in the luminal-like subtype. Ten genes, including CCNB1, CDH1, KDR, RAB25, PRKCA, etc., found which can maintain the high accordance of mRNA-protein for both breast cancer patients and cell lines in basal-like subtypes for the first time. Bi-EB provides a useful biclustering analysis tool to discover the cross patterns hidden both in multiple data matrixes (omics) and species. The implementation of the Bi-EB method in the clinical setting will have a direct impact on administrating translational research based on the cancer cell screening guidance.
Keywords: biclustering; breast cancer; multi-omics data analysis; tumor and cancer cell lines.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



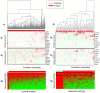
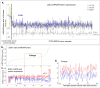
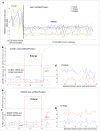
Similar articles
-
Bi-Force: large-scale bicluster editing and its application to gene expression data biclustering.Nucleic Acids Res. 2014 May;42(9):e78. doi: 10.1093/nar/gku201. Epub 2014 Mar 20. Nucleic Acids Res. 2014. PMID: 24682815 Free PMC article.
-
Identification of coherent patterns in gene expression data using an efficient biclustering algorithm and parallel coordinate visualization.BMC Bioinformatics. 2008 Apr 23;9:210. doi: 10.1186/1471-2105-9-210. BMC Bioinformatics. 2008. PMID: 18433478 Free PMC article.
-
QUBIC: a qualitative biclustering algorithm for analyses of gene expression data.Nucleic Acids Res. 2009 Aug;37(15):e101. doi: 10.1093/nar/gkp491. Epub 2009 Jun 9. Nucleic Acids Res. 2009. PMID: 19509312 Free PMC article.
-
Recent patents on biclustering algorithms for gene expression data analysis.Recent Pat DNA Gene Seq. 2011 Aug;5(2):117-25. doi: 10.2174/187221511796392097. Recent Pat DNA Gene Seq. 2011. PMID: 21529337 Review.
-
Biclustering data analysis: a comprehensive survey.Brief Bioinform. 2024 May 23;25(4):bbae342. doi: 10.1093/bib/bbae342. Brief Bioinform. 2024. PMID: 39007596 Free PMC article. Review.
References
-
- Saber H.B., Elloumi M. DNA microarray data analysis: A new survey on biclustering. Int. J. Comput. Biol. 2015;4:21–37. doi: 10.34040/IJCB.4.1.2014.36. - DOI
-
- Cheng Y., Church G.M. Biclustering of expression data. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2000;8:93–103. - PubMed
-
- Lazzeroni L., Owen A. Plaid models for gene expression data. Stat. Sin. 2002;12:61–86.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

