Deregulated Expression of Circular RNAs Is Associated with Immune Evasion and Leukemia Relapse after Allogeneic Hematopoietic Stem Cell Transplantation
- PMID: 36360223
- PMCID: PMC9689715
- DOI: 10.3390/genes13111986
Deregulated Expression of Circular RNAs Is Associated with Immune Evasion and Leukemia Relapse after Allogeneic Hematopoietic Stem Cell Transplantation
Abstract
Background: Circular RNAs (circRNAs) are a novel class of epigenetic regulators that participate in leukemogenesis. However, their roles in leukemia relapse after transplantation remain unclear.
Methods: We defined the circRNAs profile of the bone-marrow-enriched CD34+ cells from ten acute myeloid leukemia (AML) patients after transplantation (five relapse [RE] and five continuous complete remission [CR]) and four healthy controls (HCs) by RNA-seq. Differentially expressed circRNAs were validated using real-time quantitative polymerase chain reaction (RT-qPCR) in an independent cohort of six AML patients with pairwise samples at diagnosis and at relapse and six controls.
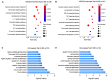
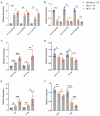
Results: The bioinformatics analysis revealed a distinct circRNAs profile in relapse patients compared with controls (CR or HCs), while there was no significant difference between CR and HCs. Functional enrichment analysis demonstrated that mRNAs co-expressed with identified circRNAs were primarily involved in immune-related pathways, including the T cell receptor signaling pathway and lymphocyte differentiation. Moreover, we performed a protein-protein interaction network based on the immune-related genes and annotated 20 hub genes. The abnormal expression of hub genes was responsible for impairing T cell co-stimulation and activation, thus contributing to the immune escape of relapse blasts. We further constructed competing endogenous RNAs (ceRNA) regulatory networks based on immune-related genes and identified 10 key circRNAs that are associated with immune evasion. Six candidate circRNAs and their associated miRNA/mRNAs in the ceRNA network were randomly selected to be validated in another set by RT-qPCR.
Conclusions: CircRNAs dysregulation may be involved in the immune evasion of relapse blasts and is associated with AML relapse. Our results identify several promising biomarkers and might provide novel insights into the biology of AML relapse post-transplantation.
Keywords: acute myeloid leukemia; allogeneic hematopoietic stem cell transplantation; circRNA; immune evasion; relapse.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Identification of Serum Exosome-Derived circRNA-miRNA-TF-mRNA Regulatory Network in Postmenopausal Osteoporosis Using Bioinformatics Analysis and Validation in Peripheral Blood-Derived Mononuclear Cells.Front Endocrinol (Lausanne). 2022 Jun 9;13:899503. doi: 10.3389/fendo.2022.899503. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35757392 Free PMC article.
-
Identification of circRNA-lncRNA-miRNA-mRNA Competitive Endogenous RNA Network as Novel Prognostic Markers for Acute Myeloid Leukemia.Genes (Basel). 2020 Jul 31;11(8):868. doi: 10.3390/genes11080868. Genes (Basel). 2020. PMID: 32751923 Free PMC article.
-
Expression profiling of N6-methyladenosine modified circRNAs in acute myeloid leukemia.Biochem Biophys Res Commun. 2022 Apr 23;601:137-145. doi: 10.1016/j.bbrc.2022.02.087. Epub 2022 Feb 23. Biochem Biophys Res Commun. 2022. PMID: 35247767
-
Role of microRNAs, circRNAs and long noncoding RNAs in acute myeloid leukemia.J Hematol Oncol. 2019 May 24;12(1):51. doi: 10.1186/s13045-019-0734-5. J Hematol Oncol. 2019. PMID: 31126316 Free PMC article. Review.
-
Epigenetic maintenance strategies after allogeneic stem cell transplantation in acute myeloid leukemia.Exp Hematol. 2022 May;109:1-10.e1. doi: 10.1016/j.exphem.2022.03.003. Epub 2022 Mar 12. Exp Hematol. 2022. PMID: 35288231 Review.
Cited by
-
Functions of Circular RNA in Human Diseases and Illnesses.Noncoding RNA. 2023 Jul 4;9(4):38. doi: 10.3390/ncrna9040038. Noncoding RNA. 2023. PMID: 37489458 Free PMC article. Review.
-
The impact of epigenetic modifications on allogeneic hematopoietic stem cell transplantation.Front Immunol. 2023 May 31;14:1188853. doi: 10.3389/fimmu.2023.1188853. eCollection 2023. Front Immunol. 2023. PMID: 37325668 Free PMC article. Review.
-
Virus-Encoded Circular RNAs: Role and Significance in Viral Infections.Int J Mol Sci. 2023 Nov 20;24(22):16547. doi: 10.3390/ijms242216547. Int J Mol Sci. 2023. PMID: 38003737 Free PMC article. Review.
References
-
- Schmid C., de Wreede L.C., van Biezen A., Finke J., Ehninger G., Ganser A., Volin L., Niederwieser D., Beelen D., Alessandrino P., et al. Outcome after relapse of myelodysplastic syndrome and secondary acute myeloid leukemia following allogeneic stem cell transplantation: A retrospective registry analysis on 698 patients by the Chronic Malignancies Working Party of the European Society of Blood and Marrow Transplantation. Haematologica. 2018;103:237–245. doi: 10.3324/haematol.2017.168716. - DOI - PMC - PubMed
-
- Gambacorta V., Beretta S., Ciccimarra M., Zito L., Giannetti K., Andrisani A., Gnani D., Zanotti L., Oliveira G., Carrabba M.G., et al. Integrated Multiomic Profiling Identifies the Epigenetic Regulator PRC2 as a Therapeutic Target to Counteract Leukemia Immune Escape and Relapse. Cancer Discov. 2022;12:1449–1461. doi: 10.1158/2159-8290.CD-21-0980. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

