3D Genome Plasticity in Normal and Diseased Neurodevelopment
- PMID: 36360237
- PMCID: PMC9690570
- DOI: 10.3390/genes13111999
3D Genome Plasticity in Normal and Diseased Neurodevelopment
Abstract
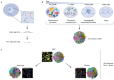
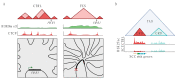
Non-random spatial organization of the chromosomal material inside the nuclei of brain cells emerges as an important regulatory layer of genome organization and function in health and disease. Here, we discuss how integrative approaches assessing chromatin in context of the 3D genome is providing new insights into normal and diseased neurodevelopment. Studies in primate (incl. human) and rodent brain have confirmed that chromosomal organization in neurons and glia undergoes highly dynamic changes during pre- and early postnatal development, with potential for plasticity across a much wider age window. For example, neuronal 3D genomes from juvenile and adult cerebral cortex and hippocampus undergo chromosomal conformation changes at hundreds of loci in the context of learning and environmental enrichment, viral infection, and neuroinflammation. Furthermore, locus-specific structural DNA variations, such as micro-deletions, duplications, repeat expansions, and retroelement insertions carry the potential to disrupt the broader epigenomic and transcriptional landscape far beyond the boundaries of the site-specific variation, highlighting the critical importance of long-range intra- and inter-chromosomal contacts for neuronal and glial function.
Keywords: 3D genome; 4D nucleome; Hi-C; brain; chromosomal conformations; cis-regulatory domain; neurodevelopment; neuropsychiatric disorder; non-coding DNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

