Seropositivity-Dependent Association between LINE-1 Methylation and Response to Methotrexate Therapy in Early Rheumatoid Arthritis Patients
- PMID: 36360249
- PMCID: PMC9690891
- DOI: 10.3390/genes13112012
Seropositivity-Dependent Association between LINE-1 Methylation and Response to Methotrexate Therapy in Early Rheumatoid Arthritis Patients
Abstract
Background: Methotrexate (MTX) is considered the first choice among disease-modifying anti-rheumatic drugs (DMARDs) for rheumatoid arthritis (RA) treatment. However, response to it varies as approximately 40% of the patients do not respond and would lose the most effective period of treatment time. Therefore, having a predictive biomarker before starting MTX treatment is of utmost importance. Methylation of long interspersed nucleotide element-1 (LINE-1) is generally considered a surrogate marker for global genomic methylation, which has been reported to associate with disease activity after MTX therapy.
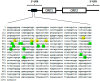
Methods: We performed a prospective study on 273 naïve early RA (ERA) patients who were treated with MTX, followed up to 12 months, and classified according to their therapy response. The baseline LINE-1 methylation levels in peripheral blood mononuclear cells (PBMC) of cases were assessed by bisulfite pyrosequencing.
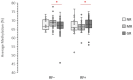
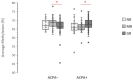
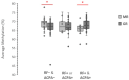
Results: Baseline LINE-1 methylation level per se turned out not to predict the response to the therapy, nor did age, sex, body mass index, or smoking status. However, if cases were stratified according to positivity to rheumatoid factor (RF) and anti-citrullinated protein antibody (ACPA) or seronegativity, we observed an opposite association between baseline LINE-1 methylation levels and optimal response to MTX therapy among responders. The best response to MTX therapy was associated with hypermethylated LINE-1 among double-positive ERA cases (p-value: 0.002) and with hypomethylated LINE-1 in seronegative ERA patients (p-value: 0.01).
Conclusion: The LINE-1 methylation level in PBMCs of naïve ERA cases associates with the degree of response to MTX therapy in an opposite way depending on the presence of RF and ACPA antibodies. Our results suggest LINE-1 methylation level as a new epigenetic biomarker for predicting the degree of response to MTX in both double-positive and seronegative ERA patients.
Keywords: DNA methylation; LINE-1; anti-citrullinated protein antibody; biomarkers; methotrexate; pharmacoepigenetics; rheumatoid arthritis; rheumatoid factor.
Conflict of interest statement
C.S. reports consulting fees from Abbvie, Roche, Lilly, Novartis, and Amgen and royalties from UpToDate. The other authors declare that they have no competing interest.
Figures

References
-
- Aletaha D., Neogi T., Silman A.J., Funovits J., Felson D.T., Bingham C.O., Birnbaum N.S., Burmester G.R., Bykerk V.P., Cohen M.D., et al. 2010 Rheumatoid Arthritis Classification Criteria: An American College of Rheumatology/European League Against Rheumatism Collaborative Initiative. Ann. Rheum. Dis. 2010;69:1580–1588. doi: 10.1136/ard.2010.138461. - DOI - PubMed
-
- Gibofsky A. Overview of Epidemiology, Pathophysiology, and Diagnosis of Rheumatoid Arthritis. Am. J. Manag. Care. 2012;18:S295–S302. - PubMed
-
- Källberg H., Ding B., Padyukov L., Bengtsson C., Rönnelid J., Klareskog L., Alfredsson L. EIRA Study Group Smoking Is a Major Preventable Risk Factor for Rheumatoid Arthritis: Estimations of Risks after Various Exposures to Cigarette Smoke. Ann. Rheum. Dis. 2011;70:508–511. doi: 10.1136/ard.2009.120899. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

