Analysis of Heat Shock Proteins Based on Amino Acids for the Tomato Genome
- PMID: 36360251
- PMCID: PMC9690137
- DOI: 10.3390/genes13112014
Analysis of Heat Shock Proteins Based on Amino Acids for the Tomato Genome
Abstract
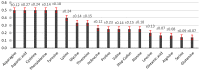
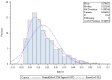
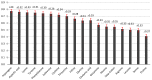
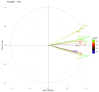
This research aimed to investigate heat shock proteins in the tomato genome through the analysis of amino acids. The highest length among sequences was found in seq19 with 3534 base pairs. This seq19 was reported and contained a family of proteins known as HsfA that have a domain of transcriptional activation for tolerance to heat and other abiotic stresses. The values of the codon adaptation index (CAI) ranged from 0.80 in Seq19 to 0.65 in Seq10, based on the mRNA of heat shock proteins for tomatoes. Asparagine (AAT, AAC), aspartic acid (GAT, GAC), phenylalanine (TTT, TTC), and tyrosine (TAT, TAC) have relative synonymous codon usage (RSCU) values bigger than 0.5. In modified relative codon bias (MRCBS), the high gene expressions of the amino acids under heat stress were histidine, tryptophan, asparagine, aspartic acid, lysine, phenylalanine, isoleucine, cysteine, and threonine. RSCU values that were less than 0.5 were considered rare codons that affected the rate of translation, and thus selection could be effective by reducing the frequency of expressed genes under heat stress. The normal distribution of RSCU shows about 68% of the values drawn from the standard normal distribution were within 0.22 and -0.22 standard deviations that tend to cluster around the mean. The most critical component based on principal component analysis (PCA) was the RSCU. These findings would help plant breeders in the development of growth habits for tomatoes during breeding programs.
Keywords: amino acids; codon adaptation index; modified relative codon bias; principal component analysis; relative synonymous codon usage.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Analysis of chromosomes and nucleotides in rice to predict gene expression through codon usage pattern.Saudi J Biol Sci. 2021 Aug;28(8):4569-4574. doi: 10.1016/j.sjbs.2021.04.059. Epub 2021 Apr 30. Saudi J Biol Sci. 2021. PMID: 34354442 Free PMC article.
-
Prediction of gene expression under drought stress in spring wheat using codon usage pattern.Saudi J Biol Sci. 2021 Jul;28(7):4000-4004. doi: 10.1016/j.sjbs.2021.04.015. Epub 2021 Apr 20. Saudi J Biol Sci. 2021. PMID: 34220257 Free PMC article.
-
The effective number of codons for individual amino acids: some codons are more optimal than others.Gene. 2003 Nov 27;320:185-90. doi: 10.1016/s0378-1119(03)00829-1. Gene. 2003. PMID: 14597402
-
Looking into the genome of Thermosynechococcus elongatus (thermophilic cyanobacteria) with codon selection and usage perspective.Int J Bioinform Res Appl. 2015;11(2):130-41. doi: 10.1504/IJBRA.2015.068088. Int J Bioinform Res Appl. 2015. PMID: 25786792
-
Differential distribution of amino acids in plants.Amino Acids. 2017 May;49(5):821-869. doi: 10.1007/s00726-017-2401-x. Epub 2017 Mar 15. Amino Acids. 2017. PMID: 28299478 Review.
Cited by
-
A Molecular Orchestration of Plant Translation under Abiotic Stress.Cells. 2023 Oct 13;12(20):2445. doi: 10.3390/cells12202445. Cells. 2023. PMID: 37887289 Free PMC article. Review.
References
-
- Araus J., Slafer G.A., Royo C., Serret M.D. Breeding for Yield Potential and Stress Adaptation in Cereals. Crit. Rev. Plant Sci. 2008;27:377–412. doi: 10.1080/07352680802467736. - DOI
-
- Hong J.C. General Aspects of Plant Transcription Factor Families. In: Gonzalez D.H., editor. Plant Transcription Factors. Academic Press; Cambridge, MA, USA: 2016. pp. 35–56. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

