Complete Chloroplast Genome Sequence of the Endemic and Endangered Plant Dendropanax oligodontus: Genome Structure, Comparative and Phylogenetic Analysis
- PMID: 36360265
- PMCID: PMC9690231
- DOI: 10.3390/genes13112028
Complete Chloroplast Genome Sequence of the Endemic and Endangered Plant Dendropanax oligodontus: Genome Structure, Comparative and Phylogenetic Analysis
Abstract
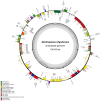
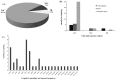
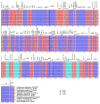
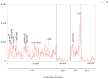
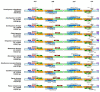
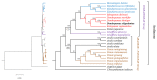
Dendropanax oligodontus, which belongs to the family Araliaceae, is an endemic and endangered species of Hainan Island, China. It has potential economic and medicinal value owing to the presence of phenylpropanoids, flavonoids, triterpenoids, etc. The analysis of the structure and characteristics of the D. oligodontus chloroplast genome (cpDNA) is crucial for understanding the genetic and phylogenetic evolution of this species. In this study, the cpDNA of D. oligodontus was sequenced for the first time using next-generation sequencing methods, assembled, and annotated. We observed a circular quadripartite structure comprising a large single-copy region (86,440 bp), a small single-copy region (18,075 bp), and a pair of inverted repeat regions (25,944 bp). The total length of the cpDNA was 156,403 bp, and the GC% was 37.99%. We found that the D. oligodontus chloroplast genome comprised 131 genes, with 86 protein-coding genes, 8 rRNA genes, and 37 tRNAs. Furthermore, we identified 26,514 codons, 13 repetitive sequences, and 43 simple sequence repeat sites in the D. oligodontus cpDNA. The most common amino acid encoded was leucine, with a strong A/T preference at the third position of the codon. The prediction of RNA editing sites in the protein-coding genes indicated that RNA editing was observed in 19 genes with a total of 54 editing sites, all of which involved C-to-T transitions. Finally, the cpDNA of 11 species of the family Araliaceae were selected for comparative analysis. The sequences of the untranslated regions and coding regions among 11 species were highly conserved, and minor differences were observed in the length of the inverted repeat regions; therefore, the cpDNAs were relatively stable and consistent among these 11 species. The variable hotspots in the genome included clpP, ycf1, rnK-rps16, rps16-trnQ, atpH-atpI, trnE-trnT, psbM-trnD, ycf3-trnS, and rpl32-trnL, providing valuable molecular markers for species authentication and regions for inferring phylogenetic relationships among them, as well as for evolutionary studies. Evolutionary selection pressure analysis indicated that the atpF gene was strongly subjected to positive environmental selection. Phylogenetic analysis indicated that D. oligodontus and Dendropanax dentiger were the most closely related species within the genus, and D. oligodontus was closely related to the genera Kalopanax and Metapanax in the Araliaceae family. Overall, the cp genomes reported in this study will provide resources for studying the genetic diversity and conservation of the endangered plant D. oligodontus, as well as resolving phylogenetic relationships within the family.
Keywords: Dendropanax; chloroplast genomes; nucleotide diversity; phylogenetic analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Complete chloroplast genome of Stephania tetrandra (Menispermaceae) from Zhejiang Province: insights into molecular structures, comparative genome analysis, mutational hotspots and phylogenetic relationships.BMC Genomics. 2021 Dec 6;22(1):880. doi: 10.1186/s12864-021-08193-x. BMC Genomics. 2021. PMID: 34872502 Free PMC article.
-
A systematic comparison of eight new plastome sequences from Ipomoea L.PeerJ. 2019 Mar 11;7:e6563. doi: 10.7717/peerj.6563. eCollection 2019. PeerJ. 2019. PMID: 30881765 Free PMC article.
-
Complete chloroplast genomes of Zingiber montanum and Zingiber zerumbet: Genome structure, comparative and phylogenetic analyses.PLoS One. 2020 Jul 31;15(7):e0236590. doi: 10.1371/journal.pone.0236590. eCollection 2020. PLoS One. 2020. PMID: 32735595 Free PMC article.
-
Dynamic evolution of the plastome in the Elm family (Ulmaceae).Planta. 2022 Dec 17;257(1):14. doi: 10.1007/s00425-022-04045-4. Planta. 2022. PMID: 36526857 Review.
-
Poaceae Chloroplast Genome Sequencing: Great Leap Forward in Recent Ten Years.Curr Genomics. 2023 Feb 14;23(6):369-384. doi: 10.2174/1389202924666221201140603. Curr Genomics. 2023. PMID: 37920556 Free PMC article. Review.
Cited by
-
Plastome structure and phylogenetic relationships of genus Hydrocotyle (apiales): provide insights into the plastome evolution of Hydrocotyle.BMC Plant Biol. 2024 Aug 15;24(1):778. doi: 10.1186/s12870-024-05483-w. BMC Plant Biol. 2024. PMID: 39148054 Free PMC article.
-
New Insights into Phylogenetic Relationship of Hydrocotyle (Araliaceae) Based on Plastid Genomes.Int J Mol Sci. 2023 Nov 22;24(23):16629. doi: 10.3390/ijms242316629. Int J Mol Sci. 2023. PMID: 38068952 Free PMC article.
References
-
- Dendropanax Decne. & Planch. Plants of the World Online. Kew Science. [(accessed on 17 August 2022)]. Available online: https://powo.science.kew.org/taxon/urn:lsid:ipni.org:names:30033431-2#so....
-
- Editorial Committee of the Flora of China. The Chinese Academy of Sciences . Flora of China. Volume 54. Science Press; Beijing, China: 1978. pp. 58–61.
-
- Giang V.N.L., Waminal N.E., Park H.S., Kim N.H., Jang W., Lee J., Yang T.J. Comprehensive comparative analysis of chloroplast genomes from seven Panax species and development of an authentication system based on species-unique single nucleotide polymorphism markers. J. Ginseng Res. 2020;44:135–144. - PMC - PubMed
-
- Wang X.M., Yang L., He J.W., Ren G., Yang M., Zhong G.Y. Research progress of chemical constituents and pharmacological activities of genus Dendropanax. Chin. J. ETMF. 2015;21:229–234.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

