Complete Mitogenomes of Ticks Ixodes acutitarsus and Ixodes ovatus Parasitizing Giant Panda: Deep Insights into the Comparative Mitogenomic and Phylogenetic Relationship of Ixodidae Species
- PMID: 36360286
- PMCID: PMC9691169
- DOI: 10.3390/genes13112049
Complete Mitogenomes of Ticks Ixodes acutitarsus and Ixodes ovatus Parasitizing Giant Panda: Deep Insights into the Comparative Mitogenomic and Phylogenetic Relationship of Ixodidae Species
Abstract
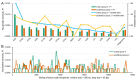
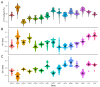
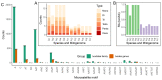
Ticks rank second in the world as vectors of disease. Tick infestation is one of the factors threatening the health and survival of giant pandas. Here, we describe the mitogenomes of Ixodes acutitarsus and Ixodes ovatus parasitizing giant pandas, and perform comparative and phylogenetic genomic analyses on the newly sequenced and other available mitogenomes of hard ticks. All six newly determined mitogenomes contain a typical gene component and share an ancient Arthropoda gene arrangement pattern. Our study suggests that I. ovatus is a species complex with high genetic divergence, indicating that different clades of I. ovatus represent distinct species. Comparative mitogenomic analyses show that the average A + T content of Ixodidae mitogenomes is 78.08%, their GC-skews are strongly negative, while AT-skews fluctuate around 0. A large number of microsatellites are detected in Ixodidae mitogenomes, and the main microsatellite motifs are mononucleotide A and trinucleotide AAT. We summarize five gene arrangement types, and identify the trnY-COX1-trnS1-COX2-trnK-ATP8-ATP6-COX3-trnG fragment is the most conserved region, whereas the region near the control region is the rearrangement hotspot in Ixodidae mitogenomes. The phylogenetic trees based on 15 genes provide a very convincing relationship (Ixodes + (Robertsicus + ((Bothriocroton + Haemaphysalis) + (Amblyomma + (Dermacentor + (Rhipicentor + (Hyalomma + Rhipicephalus))))))) with very strong supports. Remarkably, Archaeocroton sphenodonti is embedded in the Haemaphysalis clade with strong supports, resulting in paraphyly of the Haemaphysalis genus, so in-depth morphological and molecular studies are essential to determine the taxonomic status of A. sphenodonti and its closely related species. Our results provide new insights into the molecular phylogeny and evolution of hard ticks, as well as basic data for population genetics assessment and efficient surveillance and control for the giant panda-infesting ticks.
Keywords: Ixodidae; comparative mitogenomics; giant panda; mitochondrial phylogenomics; mitogenome.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures




References
-
- de Oliveira G.M.B., Araújo A.d.C., Santos J.R., da Silva I.W.G., Labruna M.B., Horta M.C. Lack of seasonality of Amblyomma rotundatum (Acari: Ixodidade) on Rhinella jimi (Anura: Bufonidae) in a semi-arid region of northeastern Brazil. Ticks Tick-Borne Dis. 2018;9:1350–1353. doi: 10.1016/j.ttbdis.2018.06.005. - DOI - PubMed
-
- Mohamed W.M.A., Moustafa M.A.M., Thu M.J., Kakisaka K., Chatanga E., Ogata S., Hayashi N., Taya Y., Ohari Y., Naguib D., et al. Comparative mitogenomics elucidates the population genetic structure of Amblyomma testudinarium in Japan and a closely related Amblyomma species in Myanmar. Evol. Appl. 2022;15:1062–1078. doi: 10.1111/eva.13426. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

