Evaluation of DNA Methylation Array for Glioma Tumor Profiling and Description of a Novel Epi-Signature to Distinguish IDH1/IDH2 Mutant and Wild-Type Tumors
- PMID: 36360312
- PMCID: PMC9690723
- DOI: 10.3390/genes13112075
Evaluation of DNA Methylation Array for Glioma Tumor Profiling and Description of a Novel Epi-Signature to Distinguish IDH1/IDH2 Mutant and Wild-Type Tumors
Abstract
Molecular biomarkers, such as IDH1/IDH2 mutations and 1p19q co-deletion, are included in the histopathological and clinical criteria currently used to diagnose and classify gliomas. IDH1/IDH2 mutation is a common feature of gliomas and is associated with a glioma-CpG island methylator phenotype (CIMP). Aberrant genomic methylation patterns can also be used to extrapolate information about copy number variation in a tumor. This project's goal was to assess the feasibility of DNA methylation array for the simultaneous detection of glioma biomarkers as a more effective testing strategy compared to existing single analyte tests.
Methods: Whole-genome methylation array (WGMA) testing was performed using 48 glioma DNA samples to detect methylation aberrations and chromosomal gains and losses. The analyzed samples include 39 tumors in the discovery cohort and 9 tumors in the replication cohort. Methylation profiles for each sample were correlated with IDH1 p.R132G mutation, immunohistochemistry (IHC), and previous 1p19q clinical testing to assess the sensitivity and specificity of the WGMA assay for the detection of these variants.
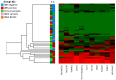
Results: We developed a DNA methylation signature to specifically distinguish a IDH1/IDH2 mutant tumor from normal samples. This signature is composed of 11 CpG sites that were significantly hypermethylated in the IDH1/IDH2 mutant group. Copy number analysis using WGMA data was able to identify five of five positive samples for 1p19q co-deletion and was concordant for all negative samples.
Conclusions: The DNA methylation signature presented here has the potential to refine the utility of WGMA to predict IDH1/IDH2 mutation status of gliomas, thus improving diagnostic yield and efficiency of laboratory testing compared to single analyte IDH1/IDH2 or 1p19q tests.
Keywords: 1p19q co-deletion; IDH1/IDH2 epi-signature; glioma; whole-genome methylation array.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Identification of retinol binding protein 1 promoter hypermethylation in isocitrate dehydrogenase 1 and 2 mutant gliomas.J Natl Cancer Inst. 2012 Oct 3;104(19):1458-69. doi: 10.1093/jnci/djs357. Epub 2012 Sep 3. J Natl Cancer Inst. 2012. PMID: 22945948 Free PMC article.
-
Isocitrate Dehydrogenase Mutations are Better Prognostic Marker than O6-methylguanine-DNA Methyltransferase Promoter Methylation in Glioblastomas - a Retrospective, Single-centre Molecular Genetics Study of Gliomas.Klin Onkol. 2017 Fall;30(5):361-371. doi: 10.14735/amko2017361. Klin Onkol. 2017. PMID: 29031038 English.
-
The comparison of clinical and biological characteristics between IDH1 and IDH2 mutations in gliomas.J Exp Clin Cancer Res. 2016 May 31;35:86. doi: 10.1186/s13046-016-0362-7. J Exp Clin Cancer Res. 2016. PMID: 27245697 Free PMC article.
-
Specific monoclonal antibodies against IDH1/2 mutations as diagnostic tools for gliomas.Brain Tumor Pathol. 2015 Jan;32(1):3-11. doi: 10.1007/s10014-014-0202-4. Epub 2014 Oct 17. Brain Tumor Pathol. 2015. PMID: 25324168 Review.
-
Molecular features assisting in diagnosis, surgery, and treatment decision making in low-grade gliomas.Neurosurg Focus. 2015 Mar;38(3):E2. doi: 10.3171/2015.1.FOCUS14745. Neurosurg Focus. 2015. PMID: 25727224 Review.
Cited by
-
Detection of diagnostic and prognostic methylation-based signatures in liquid biopsy specimens from patients with meningiomas.Nat Commun. 2023 Sep 13;14(1):5669. doi: 10.1038/s41467-023-41434-z. Nat Commun. 2023. PMID: 37704607 Free PMC article.
-
Genome-Wide Methylation Patterns in Primary Uveal Melanoma: Development of MethylSig-UM, an Epigenomic Prognostic Signature to Improve Patient Stratification.Cancers (Basel). 2024 Jul 25;16(15):2650. doi: 10.3390/cancers16152650. Cancers (Basel). 2024. PMID: 39123378 Free PMC article.
References
-
- Ostrom Q.T., Cioffi G., Gittleman H., Patil N., Waite K., Kruchko C., Barnholtz-Sloan J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2012–2016. Neuro Oncol. 2019;21((Suppl. 5)):v1–v100. doi: 10.1093/neuonc/noz150. - DOI - PMC - PubMed
-
- Louis D.N., Perry A., Reifenberger G., Von Deimling A., Figarella-Branger D., Cavenee W.K., Ohgaki H., Wiestler O.D., Kleihues P., Ellison D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. - DOI - PubMed
-
- Louis D.N., Perry A., Wesseling P., Brat D.J., Cree I.A., Figarella-Branger D., Hawkins C., Ng H.K., Pfister S.M., Reifenberger G., et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021;23:1231–1251. doi: 10.1093/neuonc/noab106. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

