Evaluating EcxR for Its Possible Role in Ehrlichia chaffeensis Gene Regulation
- PMID: 36361509
- PMCID: PMC9657007
- DOI: 10.3390/ijms232112719
Evaluating EcxR for Its Possible Role in Ehrlichia chaffeensis Gene Regulation
Abstract
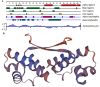
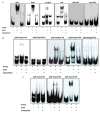
Ehrlichia chaffeensis, a tick-transmitted intraphagosomal bacterium, is the causative agent of human monocytic ehrlichiosis. The pathogen also infects several other vertebrate hosts. E. chaffeensis has a biphasic developmental cycle during its growth in vertebrate monocytes/macrophages and invertebrate tick cells. Host- and vector-specific differences in the gene expression from many genes of E. chaffeensis are well documented. It is unclear how the organism regulates gene expression during its developmental cycle and for its adaptation to vertebrate and tick host cell environments. We previously mapped promoters of several E. chaffeensis genes which are recognized by its only two sigma factors: σ32 and σ70. In the current study, we investigated in assessing five predicted E. chaffeensis transcription regulators; EcxR, CtrA, MerR, HU and Tr1 for their possible roles in regulating the pathogen gene expression. Promoter segments of three genes each transcribed with the RNA polymerase containing σ70 (HU, P28-Omp14 and P28-Omp19) and σ32 (ClpB, DnaK and GroES/L) were evaluated by employing multiple independent molecular methods. We report that EcxR binds to all six promoters tested. Promoter-specific binding of EcxR to several gene promoters results in varying levels of gene expression enhancement. This is the first detailed molecular characterization of transcription regulators where we identified EcxR as a gene regulator having multiple promoter-specific interactions.
Keywords: ehrlichia tick-borne disease; obligate intracellular bacteria; rickettsial gene regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Ganguly S., Mukhopadhayay S.K. Tick-borne ehrlichiosis infection in human beings. J. Vector Borne. Dis. 2008;45:273–280. - PubMed
-
- Dawson J.E., Biggie K.L., Warner C.K., Cookson K., Jenkins S., Levine J.F., Olson J.G. Polymerase chain reaction evidence of Ehrlichia chaffeensis, an etiologic agent of human monocytic ehrlichiosis, in dogs, from southeast Virginia. Am. J. Vet. Res. 1996;57:1175–1179. - PubMed
-
- Lockhart J.M., Davidson W.R., Stallknecht D.E., Dawson J.E., Howerth E.W. Isolation of Ehrlichia chaffeensis from wild white-tailed deer (Odocoileus virginianus) confirms their role as natural reservoir hosts. J. Clin. Microbiol. 1997;35:1681–1686. doi: 10.1128/jcm.35.7.1681-1686.1997. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

