Genome-Wide Identification and Expression Analysis of Senescence-Associated Genes in Grapevine (Vitis vinifera L.) Reveal Their Potential Functions in Leaf Senescence Order
- PMID: 36361520
- PMCID: PMC9656468
- DOI: 10.3390/ijms232112731
Genome-Wide Identification and Expression Analysis of Senescence-Associated Genes in Grapevine (Vitis vinifera L.) Reveal Their Potential Functions in Leaf Senescence Order
Abstract
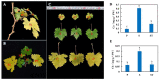
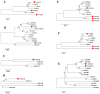
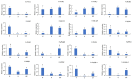
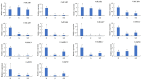
Natural leaf senescence is an acclimation strategy that enables plants to reallocate nutrients. In the present study, interestingly, we found that the basal mature leaves of grapevine primary shoots (P) exhibited the earliest senescence, followed by the apical young leaves of secondary shoots (ST), and then the basal mature leaves of secondary shoots (S). The Chl level decreased with the extent of leaf senescence. According to the genome-wide identification and expression analysis, sixteen senescence-associated genes (SAGs) involved in Chl breakdown were identified in the grapevine genome. Their expression patterns showed that the transcript changes in VvSGR, VvPPH2, and VvFtsH6-2 corresponded to the changes in Chl content among P, S, and ST. The changes in the transcription of VvNYC1, VvSGR, VvPAO1, VvPAO2, VvPAO4, VvPPH1, VvPPH3, and VvFtsH6-1 only contributed to low Chl levels in P. The cis-element analysis indicated that these SAGs possessed several light- and hormone-responsive elements in their promoters. Among them, ABA-responsive elements were found in twelve of the sixteen promoters of SAGs. Correspondingly, ABA-signaling components presented various changes in transcription among P, S, and ST. The transcription changes in VvbZIP45 and VvSnRK2.1 were similar to those in VvSGR, VvPPH2, and VvFtsH6-2. The other nine ABA-signaling components, which included VvRCAR2, VvRCAR4, VvRCAR6, VvRCAR7, VvRCAR2, VvPP2C4, VvPP2C9, VvbZIP25, and VvSnRK2.3, were highly expressed in P but there was no difference between S and ST, with similar expression patterns for VvNYC1, VvSGR, VvPAO1, VvPAO2, VvPAO4, VvPPH1, VvPPH3, and VvFtsH6-1. These results suggested that the senescence of P and ST could be regulated by different members of Chl breakdown-related SAGs and ABA-signaling components. These findings provide us with important candidate genes to further study the regulation mechanism of leaf senescence order in grapevine.
Keywords: ABA; chlorophyll; gene expression; grapevine; leaf order; senescence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Noodén L.D. Plant Cell Death Processes. Academic Press; London, UK: 2004.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

