SLU7: A New Hub of Gene Expression Regulation-From Epigenetics to Protein Stability in Health and Disease
- PMID: 36362191
- PMCID: PMC9658179
- DOI: 10.3390/ijms232113411
SLU7: A New Hub of Gene Expression Regulation-From Epigenetics to Protein Stability in Health and Disease
Abstract
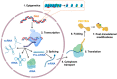
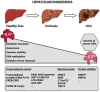
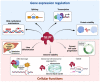
SLU7 (Splicing factor synergistic lethal with U5 snRNA 7) was first identified as a splicing factor necessary for the correct selection of 3' splice sites, strongly impacting on the diversity of gene transcripts in a cell. More recent studies have uncovered new and non-redundant roles of SLU7 as an integrative hub of different levels of gene expression regulation, including epigenetic DNA remodeling, modulation of transcription and protein stability. Here we review those findings, the multiple factors and mechanisms implicated as well as the cellular functions affected. For instance, SLU7 is essential to secure liver differentiation, genome integrity acting at different levels and a correct cell cycle progression. Accordingly, the aberrant expression of SLU7 could be associated with human diseases including cancer, although strikingly, it is an essential survival factor for cancer cells. Finally, we discuss the implications of SLU7 in pathophysiology, with particular emphasis on the progression of liver disease and its possible role as a therapeutic target in human cancer.
Keywords: alternative splicing; cancer; cell cycle; differentiation; epigenetics; gene expression; genomic stability; liver pathophysiology; stress; transcription.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Uzman A. In: Molecular Biology of the Cell. 4th ed.; Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P., editors. Garland Science; New York, NY, USA: 2003. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

