Selection of Diagnostically Significant Regions of the SLC26A4 Gene Involved in Hearing Loss
- PMID: 36362242
- PMCID: PMC9655724
- DOI: 10.3390/ijms232113453
Selection of Diagnostically Significant Regions of the SLC26A4 Gene Involved in Hearing Loss
Abstract
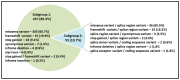
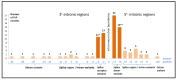
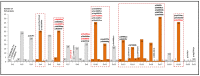
Screening pathogenic variants in the SLC26A4 gene is an important part of molecular genetic testing for hearing loss (HL) since they are one of the common causes of hereditary HL in many populations. However, a large size of the SLC26A4 gene (20 coding exons) predetermines the difficulties of its complete mutational analysis, especially in large samples of patients. In addition, the regional or ethno-specific prevalence of SLC26A4 pathogenic variants has not yet been fully elucidated, except variants c.919-2A>G and c.2168A>G (p.His723Arg), which have been proven to be most common in Asian populations. We explored the distribution of currently known pathogenic and likely pathogenic (PLP) variants across the SLC26A4 gene sequence presented in the Deafness Variation Database for the selection of potential diagnostically important parts of this gene. As a result of this bioinformatic analysis, we found that molecular testing ten SLC26A4 exons (4, 6, 10, 11, 13−17 and 19) with flanking intronic regions can provide a diagnostic rate of 61.9% for all PLP variants in the SLC26A4 gene. The primary sequencing of these SLC26A4 regions may be applied as an initial effective diagnostic testing in samples of patients of unknown ethnicity or as a subsequent step after the targeted testing of already-known ethno- or region-specific pathogenic SLC26A4 variants.
Keywords: SLC26A4; bioinformatic analysis; hearing loss; molecular testing; pathogenic variants; pendrin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- World Health Organization. [(accessed on 1 June 2022)]. Available online: https://www.who.int/en/news-room/fact-sheets/detail/deafness-and-hearing....
-
- Van Camp G., Smith R.J.H. Hereditary Hearing Loss Homepage. [(accessed on 1 June 2022)]. Available online: https://hereditaryhearingloss.org.
-
- Park H.J., Shaukat S., Liu X.Z., Hahn S.H., Naz S., Ghosh M., Kim H.N., Moon S.K., Abe S., Tukamoto K., et al. Origins and frequencies of SLC26A4 (PDS) mutations in east and south Asians: Global implications for the epidemiology of deafness. J. Med. Genet. 2003;40:242–248. doi: 10.1136/jmg.40.4.242. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

