Vulnerable Atherosclerotic Plaque: Is There a Molecular Signature?
- PMID: 36362423
- PMCID: PMC9656166
- DOI: 10.3390/ijms232113638
Vulnerable Atherosclerotic Plaque: Is There a Molecular Signature?
Abstract
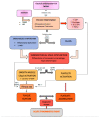
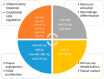
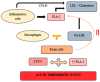
Atherosclerosis and its clinical manifestations, coronary and cerebral artery diseases, are the most common cause of death worldwide. The main pathophysiological mechanism for these complications is the rupture of vulnerable atherosclerotic plaques and subsequent thrombosis. Pathological studies of the vulnerable lesions showed that more frequently, plaques rich in lipids and with a high level of inflammation, responsible for mild or moderate stenosis, are more prone to rupture, leading to acute events. Identifying the vulnerable plaques helps to stratify patients at risk of developing acute vascular events. Traditional imaging methods based on plaque appearance and size are not reliable in prediction the risk of rupture. Intravascular imaging is a novel technique able to identify vulnerable lesions, but it is invasive and an operator-dependent technique. This review aims to summarize the current data from literature regarding the main biomarkers involved in the attempt to diagnose vulnerable atherosclerotic lesions. These biomarkers could be the base for risk stratification and development of the new therapeutic drugs in the treatment of patients with vulnerable atherosclerotic plaques.
Keywords: atherosclerosis; inflammatory biomarkers; micro-RNAs; oxidative stress; proteomics; vulnerable plaque.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Chiorescu R.M., Mocan M., Barta A., Stoia M.A., Anton P., Mocan-Hognogi D.L., Cerasela M., Goidescu A.D.F. Hypoalbuminemia—Prognostic factor for the short-term evolution of patients with unstable angina. Hum. Vet. Med. 2017;11:160–165.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

