Stieleria sedimenti sp. nov., a Novel Member of the Family Pirellulaceae with Antimicrobial Activity Isolated in Portugal from Brackish Sediments
- PMID: 36363743
- PMCID: PMC9692418
- DOI: 10.3390/microorganisms10112151
Stieleria sedimenti sp. nov., a Novel Member of the Family Pirellulaceae with Antimicrobial Activity Isolated in Portugal from Brackish Sediments
Abstract
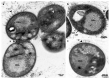
The phylum Planctomycetota is known for having uncommon biological features. Recently, biotechnological applications of its members have started to be explored, namely in the genus Stieleria. Here, we formally describe a novel Stieleriaisolate designated as strain ICT_E10.1T, obtained from sediments collected in the Tagus estuary (Portugal). Strain ICT_E10.1T is pink-pigmented, spherical to ovoid in shape, and 1.7 µm ± 0.3 × 1.4 µm ± 0.3 in size. Cells cluster strongly in aggregates or small chains, divide by budding, and have prominent fimbriae. Strain ICT_E10.1T is heterotrophic and aerobic. Growth occurs from 20 to 30 °C, from 0.5 to 3% (w/v) NaCl, and from pH 6.5 to 11.0. The analysis of the 16S rRNA gene sequence placed strain ICT_E10.1T into the genus Stieleria with Stieleria neptunia Enr13T as the closest validly described relative. The genome size is 9,813,311 bp and the DNA G+C content is 58.8 mol%. Morphological, physiological, and genomic analyses support the separation of this strain into a novel species, for which we propose the name Stieleria sedimenti represented by strain ICT_E10.1T as the type of strain (=CECT 30514T= DSM 113784T). Furthermore, this isolate showed biotechnological potential by displaying relevant biosynthetic gene clusters and potent activity against Staphylococcus aureus.
Keywords: Alcochete; Planctomycetota; bioactivity screenings; biosynthetic gene clusters; carbohydrate-active enzymes; iChip; novel taxa; organic extraction; transmission electron microscopy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
Grants and funding
- SFRH/BD/145577/2019/Fundação para a Ciência e Tecnologia
- FCT UIDB/04423/2020/Fundação para a Ciência e Tecnologia
- UIDP/04423/2020/Fundação para a Ciência e Tecnologia
- NORTE-01-0145-FEDER-000040/Norte Portugal Regional Operational Program (NORTE 2020)
- AFR scheme, ref. 14583934/National Research Fund of Luxembourg
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

