Comprehensive Phenotyping in Inflammatory Bowel Disease: Search for Biomarker Algorithms in the Transkingdom Interactions Context
- PMID: 36363782
- PMCID: PMC9698371
- DOI: 10.3390/microorganisms10112190
Comprehensive Phenotyping in Inflammatory Bowel Disease: Search for Biomarker Algorithms in the Transkingdom Interactions Context
Abstract
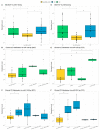
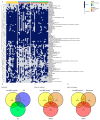
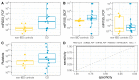
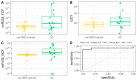
Inflammatory bowel disease (IBD) is the most common form of intestinal inflammation associated with a dysregulated immune system response to the commensal microbiota in a genetically susceptible host. IBD includes ulcerative colitis (UC) and Crohn's disease (CD), both of which are remarkably heterogeneous in their clinical presentation and response to treatment. This translates into a notable diagnostic challenge, especially in underdeveloped countries where IBD is on the rise and access to diagnosis or treatment is not always accessible for chronic diseases. The present work characterized, for the first time in our region, epigenetic biomarkers and gut microbial profiles associated with UC and CD patients in the Buenos Aires Metropolitan area and revealed differences between non-IBD controls and IBD patients. General metabolic functions associated with the gut microbiota, as well as core microorganisms within groups, were also analyzed. Additionally, the gut microbiota analysis was integrated with relevant clinical, biochemical and epigenetic markers considered in the follow-up of patients with IBD, with the aim of generating more powerful diagnostic tools to discriminate phenotypes. Overall, our study provides new insights into data analysis algorithms to promote comprehensive phenotyping tools using quantitative and qualitative analysis in a transkingdom interactions network context.
Keywords: comprehensive-phenotyping; crohn-disease; gut-microbiota; ulcerative-colitis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Magro F., Gionchetti P., Eliakim R., Ardizzone S., Armuzzi A., Barreiro-de Acosta M., Burisch J., Gecse K.B., Hart A.L., Hindryckx P., et al. Third European Evidence-based Consensus on Diagnosis and Management of Ulcerative Colitis. Part 1: Definitions, Diagnosis, Extra-intestinal Manifestations, Pregnancy, Cancer Surveillance, Surgery, and Ileo-anal Pouch Disorders. J. Crohn’s Colitis. 2017;11:649–670. doi: 10.1093/ecco-jcc/jjx008. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

