The Role of Hidden Conformers in Determination of Conformational Preferences of Mefenamic Acid by NOESY Spectroscopy
- PMID: 36365095
- PMCID: PMC9696638
- DOI: 10.3390/pharmaceutics14112276
The Role of Hidden Conformers in Determination of Conformational Preferences of Mefenamic Acid by NOESY Spectroscopy
Abstract
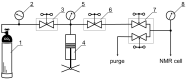
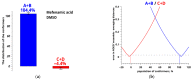
Mefenamic acid has been used as a non-steroidal anti-inflammatory drug for a long time. However, its practical use is quite limited due to a number of side effects on the intestinal organs. Conformational polymorphism provides mefenamic acid with unique properties regarding possible modifications obtained during the micronization process, which can improve pharmacokinetics and minimize side effects. Micronization can be performed by decompression of supercritical fluids; methods such as rapid expansion of the supercritical solution have proven their efficiency. However, this group of methods is poorly applicable for compounds with low solubility, and the modification of the method using a pharmaceutically suitable co-solvent may be useful. In our case, addition of only 2 mol% dimethyl sulfoxide increased the solubility remarkably. Information on the conformational state may be critically important for carrying out micronization. In this work, structural analysis and estimate of conformational preferences of mefenamic acid in dimethyl sulfoxide-d6 (at 25 °C and 0.1 MPa) and in a mixed solvent supercritical carbon dioxide + dimethyl sulfoxide-d6 (45 °C, 9 MPa) were performed based on nuclear Overhauser effect spectroscopy. Results show changes in the conformation fractions depending on the medium used. The importance of allowing for hidden conformers in estimating the conformational state was demonstrated in the analysis. Obtained results may be useful for improving micronization parameters.
Keywords: fenamates; high-pressure NMR; spatial structure; supercritical fluid.
Conflict of interest statement
The authors declare no conflict of interest.
Figures









References
-
- Harbut M.B., Vilchèze C., Luo X., Hensler M.E., Guo H., Yang B., Chatterjee A.K., Nizet V., Jacobs W.R., Schultz P.G., et al. Auranofin Exerts Broad-Spectrum Bactericidal Activities by Targeting Thiol-Redox Homeostasis. Proc. Natl. Acad. Sci. USA. 2015;112:4453–4458. doi: 10.1073/pnas.1504022112. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

