The Changes of Microbial Communities and Key Metabolites after Early Bursaphelenchus xylophilus Invasion of Pinus massoniana
- PMID: 36365304
- PMCID: PMC9653782
- DOI: 10.3390/plants11212849
The Changes of Microbial Communities and Key Metabolites after Early Bursaphelenchus xylophilus Invasion of Pinus massoniana
Abstract
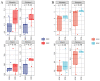
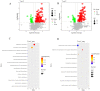
Pine wood nematode, Bursaphelenchus xylophilus, is a worldwide pest of pine trees, spreading at an alarming rate and with great ecological adaptability. In the process of causing disease, the nematode causes metabolic disorders and changes in the endophytic microbial community of the pine tree. However, the changes at the pine nidus during early nematode invasion have not been well studied, especially the differential metabolites, in Pinus massoniana, the main host of B. xylophilus in China. In this study, we analyzed the endophytic bacterial and fungal communities associated with healthy and B. xylophilus-caused wilted pine trees. The results show that 1333 bacterial OTUs and 502 fungal OTUs were annotated from P. massoniana stem samples. The abundance of bacterial communities in pine trees varies more following infection by B. xylophilus, but the abundance changes of fungal communities are less visible. There were significant differences in endophytic microbial diversity between wilted and healthy P. massoniana. In wilted pine trees, Actinobacteria and Bacteroidia were differential indicators of bacterial communities, whereas, in healthy pine trees, Rhizobiales in the Proteobacteria phylum were the major markers of bacterial communities. Meanwhile, the differential markers of fungal communities in healthy pines are Malasseziales, Tremellales, Sordariales, and Fusarium, whereas Pleosporaceae is the key marker of fungal communities in wilted pines. Our study examines the effect of changes in the endophytic microbial community on the health of pine trees that may be caused by B. xylophilus infection. In parallel, a non-targeted metabolomic study based on liquid mass spectrometry (LC-MS) technology was conducted on pine trees inoculated with pine nematodes and healthy pine trees with a view to identifying key compounds affecting early pine lesions. Ultimately, 307 distinctly different metabolites were identified. Among them, the riboflavin metabolic pathway in pine trees may play a key role in the early pathogenesis of pine wood nematode disease.
Keywords: endophytic microbes; metabolite profiling; microbial community; pines; pinewood nematode.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures





References
-
- Vicente C., Espada M., Vieira P., Mota M. Pine Wilt Disease: A threat to European forestry. Eur. J. Plant Pathol. 2012;133:89–99. doi: 10.1007/s10658-011-9924-x. - DOI
-
- Kojima K., Kamijyo A., Masumori M., Sasaki S. Cellulase activities of pine-wood nematode isolates with different virulences. J. Jpn. For. Soc. 1994;76:258–262.
-
- Odani K., Sasaki S., Nishiyama Y., Yamamoto N. Early Symptom Development of the Pine Wilt Disease by Hydrolytic Enzymes Produced by the Pine Wood Nematodes. Drug Saf. 2008;38:1032.
-
- Kuroda K. The mechanism oftra—Cheid cavitation in trees infected with wilt diseases. Proc. IUFRO Work. Party. 2002;7:17–23.
Grants and funding
LinkOut - more resources
Full Text Sources

