Salt Stress Tolerance in Casuarina glauca: Insights from the Branchlets Transcriptome
- PMID: 36365395
- PMCID: PMC9658546
- DOI: 10.3390/plants11212942
Salt Stress Tolerance in Casuarina glauca: Insights from the Branchlets Transcriptome
Abstract
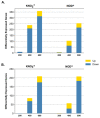
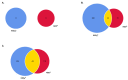
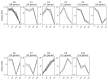
Climate change and the accelerated rate of population growth are imposing a progressive degradation of natural ecosystems worldwide. In this context, the use of pioneer trees represents a powerful approach to reverse the situation. Among others, N2-fixing actinorhizal trees constitute important elements of plant communities and have been successfully used in land reclamation at a global scale. In this study, we have analyzed the transcriptome of the photosynthetic organs of Casuarina glauca (branchlets) to unravel the molecular mechanisms underlying salt stress tolerance. For that, C. glauca plants supplied either with chemical nitrogen (KNO3+) or nodulated by Frankia (NOD+) were exposed to a gradient of salt concentrations (200, 400, and 600 mM NaCl) and RNA-Seq was performed. An average of ca. 25 million clean reads was obtained for each group of plants, corresponding to 86,202 unigenes. The patterns of differentially expressed genes (DEGs) clearly separate two groups: (i) control- and 200 mM NaCl-treated plants, and (ii) 400 and 600 mM NaCl-treated plants. Additionally, although the number of total transcripts was relatively high in both plant groups, the percentage of significant DEGs was very low, ranging from 6 (200 mM NaCl/NOD+) to 314 (600 mM NaCl/KNO3+), mostly involving down-regulation. The vast majority of up-regulated genes was related to regulatory processes, reinforcing the hypothesis that some ecotypes of C. glauca have a strong stress-responsive system with an extensive set of constitutive defense mechanisms, complemented by a tight mechanism of transcriptional and post-transcriptional regulation. The results suggest that the robustness of the stress response system in C. glauca is regulated by a limited number of genes that tightly regulate detoxification and protein/enzyme stability, highlighting the complexity of the molecular interactions leading to salinity tolerance in this species.
Keywords: Casuarina glauca; Frankia; Illumina RNA-Seq; actinorhizal plants; salt-tolerance.
Conflict of interest statement
The authors declare that there is no potential conflict of interest.
Figures



References
-
- Lapenis A. A 50-Year-Old Global Warming Forecast That Still Holds Up. Eos. 2020;101:1–10. doi: 10.1029/2020EO151822. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

