Metabolic Responses and Pathway Changes of Vero Cells under High-Vitamin B Medium
- PMID: 36366296
- PMCID: PMC9693426
- DOI: 10.3390/vaccines10111787
Metabolic Responses and Pathway Changes of Vero Cells under High-Vitamin B Medium
Abstract
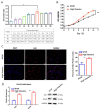
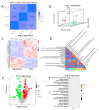
The production efficiency of a cell substrate directly affects the yield of target products such as viruses, while its density is mainly regulated by the type of culture medium and culture conditions. In this study, Vero cells were used as model cells for systematic medium screening, and a high-efficiency medium for biological drug production was identified. Through the results of cell proliferation by a cell counting kit (CCK)-8 assay, 5-Ethynyl-2'-deoxyuridine(EdU) assay, real-time quantitative PCR (RT-qPCR) and Western blotting, we found that adding an appropriate amount of vitamin B to the conventional basic medium can significantly improve and maintain the high-density growth of Vero cells. In addition, the molecular mechanism of the high-density culture of Vero cells promoted by B vitamins is explained for the first time by using the systems multi-omics analysis methods. Here, we determined that B vitamins regulate cell proliferation through the synthesis and metabolism of unsaturated fatty acids, affecting the productivity of cell substrate in industrial production. This study provides an important tool for the screening of key components of cell-based high-efficiency medium.
Keywords: Vero cell; culture medium; multi-omics analysis; vitamin B.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Kinetic characterization of vero cell metabolism in a serum-free batch culture process.Biotechnol Bioeng. 2010 Sep 1;107(1):143-53. doi: 10.1002/bit.22783. Biotechnol Bioeng. 2010. PMID: 20506276
-
A microcarrier cell culture process for propagating rabies virus in Vero cells grown in a stirred bioreactor under fully animal component free conditions.Vaccine. 2007 May 10;25(19):3879-89. doi: 10.1016/j.vaccine.2007.01.086. Epub 2007 Feb 5. Vaccine. 2007. PMID: 17307281
-
Effects of hsa_circ_0000711 expression level on proliferation and apoptosis of hepatoma cells.Eur Rev Med Pharmacol Sci. 2020 Apr;24(8):4161-4171. doi: 10.26355/eurrev_202004_20996. Eur Rev Med Pharmacol Sci. 2020. PMID: 32373952
-
Culture media for propagation of mammalian cells, viruses, and other biologicals.Adv Biotechnol Processes. 1985;5:1-30. Adv Biotechnol Processes. 1985. PMID: 2417609 Review.
-
Cooperation of liver cells in health and disease.Adv Anat Embryol Cell Biol. 2001;161:III-XIII, 1-151. doi: 10.1007/978-3-642-56553-3. Adv Anat Embryol Cell Biol. 2001. PMID: 11729749 Review.
Cited by
-
Secretomic changes of amyloid beta peptides on Alzheimer's disease related proteins in differentiated human SH-SY5Y neuroblastoma cells.PeerJ. 2024 Jul 17;12:e17732. doi: 10.7717/peerj.17732. eCollection 2024. PeerJ. 2024. PMID: 39035166 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

