Evolutionary Trajectories of Avian Avulaviruses and Vaccines Compatibilities in Poultry
- PMID: 36366369
- PMCID: PMC9698863
- DOI: 10.3390/vaccines10111862
Evolutionary Trajectories of Avian Avulaviruses and Vaccines Compatibilities in Poultry
Abstract
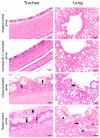
Newcastle disease virus (NDV) causes one of the highly infectious avian diseases in poultry leading to genuine financial misfortunes around the world. Recently, there has been an increasing trend in the number of ND-associated outbreaks in commercial Jordanian poultry flocks indicating a possible complex evolutionary dynamic of NDV infections in the country. To underpin the dynamics of circulating NDV strains and to assess the vaccine-escape potential, a total of 130 samples were collected from different poultry flocks in six Jordanian Governorates during 2019-2021. Twenty positive isolates, based on real-time reverse transcriptase PCR, were used for further genetic characterization and evolutionary analysis. Our results showed that there is a high evolutionary distance between the newly identified NDV strains (genotype VII.1.1) in this study and the commercially used vaccines (genotypes I and II), suggesting that circulating NDV field strains are under constant evolutionary pressure. These mutations may significantly affect flocks that have received vaccinations as well as flocks with insufficient immunity in terms of viral immunity and disease dynamics. To assess this further, we investigated the efficacy of the heterologous inactivated LaSota or homologous genotype VII.1.1 vaccine for their protection against virulent NDV in chicken. Vaccine-induced immunity was evaluated based on the serology, and protection efficacy was assessed based on clinical signs, survival rates, histopathology, and viral shedding. Chickens vaccinated with the inactivated genotype VII.1.1 based vaccine showed 100% protection with a significant reduction in virus shedding, and ameliorated histopathology lesions compared to LaSota vaccinated chicks that showed 60% protection. These results revealed that the usage of NDV inactivated vaccine from the circulating field strains can successfully ameliorate the clinical outcome and virus pathobiology in vaccinated chicks and will serve as an effective vaccine against the threat posed by commonly circulating NDV strains in the poultry industry.
Keywords: Jordan; avian orthoavulaviruses 1; efficacy; evolutionary pressure; vaccine.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
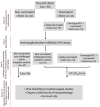


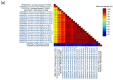


References
-
- Lamb R.A., Parks G.D. Paramyxoviridae: The viruses and their replication. In: Knipe D.M., Howley P.M., editors. Fields Virology. 5th ed. Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2006. pp. 1449–1496.
LinkOut - more resources
Full Text Sources

