Computational Design of a Chimeric Vaccine against Plesiomonas shigelloides Using Pan-Genome and Reverse Vaccinology
- PMID: 36366394
- PMCID: PMC9697808
- DOI: 10.3390/vaccines10111886
Computational Design of a Chimeric Vaccine against Plesiomonas shigelloides Using Pan-Genome and Reverse Vaccinology
Abstract
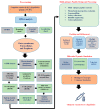
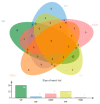
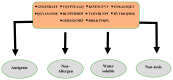
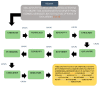
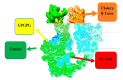
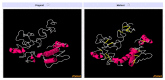
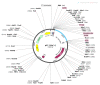
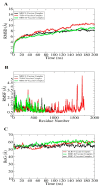
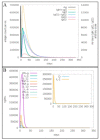
The swift emergence of antibiotic resistance (AR) in bacterial pathogens to make themselves adaptable to changing environments has become an alarming health issue. To prevent AR infection, many ways can be accomplished such as by decreasing the misuse of antibiotics in human and animal medicine. Among these AR bacterial species, Plesiomonas shigelloides is one of the etiological agents of intestinal infection in humans. It is a gram-negative rod-shaped bacterium that is highly resistant to several classes of antibiotics, and no licensed vaccine against the aforementioned pathogen is available. Hence, substantial efforts are required to screen protective antigens from the pathogen whole genome that can be subjected easily to experimental evaluations. Here, we employed a reverse vaccinology (RV) approach to design a multi-antigenic epitopes based vaccine against P. shigelloides. The complete genomes of P. shigelloides were retrieved from the National Center for Biotechnological Information (NCBI) that on average consist of 5226 proteins. The complete proteomes were subjected to different subtractive proteomics filters, and in the results of that analysis, out of total proteins, 2399 were revealed as non-redundant and 2827 as redundant proteins. The non-redundant proteins were further checked for subcellular localization analysis, in which three were localized in the extracellular matrix, eight were outer membrane, and 13 were found in the periplasmic membrane. All surface localized proteins were found to be virulent. Out of a total of 24 virulent proteins, three proteins (flagellar hook protein (FlgE), hypothetical protein, and TonB-dependent hemoglobin/transferrin/lactoferrin family receptor protein) were considered as potential vaccine targets and subjected to epitopes prediction. The predicted epitopes were further examined for antigenicity, toxicity, and solubility. A total of 10 epitopes were selected (GFKESRAEF, VQVPTEAGQ, KINENGVVV, ENKALSQET, QGYASANDE, RLNPTDSRW, TLDYRLNPT, RVTKKQSDK, GEREGKNRP, RDKKTNQPL). The selected epitopes were linked with each other via specific GPGPG linkers in order to design a multi-epitopes vaccine construct, and linked with cholera toxin B subunit adjuvant to make the designed vaccine construct more efficient in terms of antigenicity. The 3D structure of the vaccine construct was modeled ab initio as no appropriate template was available. Furthermore, molecular docking was carried out to check the interaction affinity of the designed vaccine with major histocompatibility complex (MHC-)I (PDB ID: 1L1Y), MHC-II (1KG0), and toll-like receptor 4 ((TLR-4) (PDB: 4G8A). Molecular dynamic simulation was applied to evaluate the dynamic behavior of vaccine-receptor complexes. Lastly, the binding free energies of the vaccine with receptors were estimated by using MMPB/GBSA methods. All of the aforementioned analyses concluded that the designed vaccine molecule as a good candidate to be used in experimental studies to disclose its immune protective efficacy in animal models.
Keywords: Plesiomonas shigelloides; immunoinformatics; molecular docking; multi-epitopes.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Caniça M., Manageiro V., Abriouel H., Moran-Gilad J., Franz C.M.A.P. Antibiotic resistance in foodborne bacteria. Trends Food Sci. Technol. 2019;84:41–44. doi: 10.1016/j.tifs.2018.08.001. - DOI
-
- PCAST . National Action Plan for Combatting Antibiotic-Resistant Bacteria. White House; Washington, DC, USA: 2015.
LinkOut - more resources
Full Text Sources
Research Materials

