Cocktail, a Computer Program for Modelling Bacteriophage Infection Kinetics
- PMID: 36366581
- PMCID: PMC9695944
- DOI: 10.3390/v14112483
Cocktail, a Computer Program for Modelling Bacteriophage Infection Kinetics
Abstract
Cocktail is an easy-to-use computer program for mathematical modelling of bacteriophage (phage) infection kinetics in a chemostat. The infection of bacteria by phages results in complicated dynamic processes as both have the ability to multiply and change during the course of an infection. There is a need for a simple way to visualise these processes, not least due to the increased interest in phage therapy. Cocktail is completely self-contained and runs on a Windows 64-bit operating system. By changing the publicly available source code, the program can be developed in the directions that users see fit. Cocktail's models consist of coupled differential equations that describe the infection of a bacterium in a vessel by one or two (interfering) phages. In the models, the bacterial population can be controlled by sixteen parameters, for example, through different growth rates, phage resistance, metabolically inactive cells or biofilm formation. The phages can be controlled by eight parameters each, such as different adsorption rates or latency periods. As the models in Cocktail describe the infection kinetics of phages in vitro, the program is primarily intended to generate hypotheses, but the results can however be indicative in the application of phage therapy.
Keywords: bacteriophage; computer program; infection kinetics; mathematical modelling; phage therapy.
Conflict of interest statement
The author declares no conflict of interest.
Figures
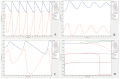
References
-
- Międzybrodzki R., Borysowski J., Weber-Dąbrowska B., Fortuna W., Letkiewicz S., Szufnarowski K., Pawełczyk Z., Rogóż P., Kłak M., Wojtasik E.E., et al. Clinical Aspects of Phage Therapy. Adv. Virus Res. 2012;83:73–121. - PubMed
-
- Chanishvili N. Phage Therapy-History from Twort and d’Herelle Through Soviet Experience to Current Approaches. Adv. Virus Res. 2012;83:3–40. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

