Adaptation in Outbred Sexual Yeast is Repeatable, Polygenic and Favors Rare Haplotypes
- PMID: 36366952
- PMCID: PMC9728589
- DOI: 10.1093/molbev/msac248
Adaptation in Outbred Sexual Yeast is Repeatable, Polygenic and Favors Rare Haplotypes
Abstract
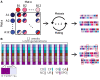
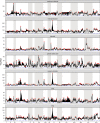
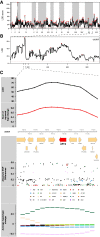
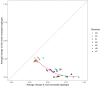
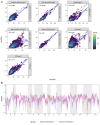
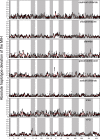
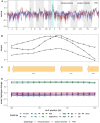
We carried out a 200 generation Evolve and Resequence (E&R) experiment initiated from an outbred diploid recombined 18-way synthetic base population. Replicate populations were evolved at large effective population sizes (>105 individuals), exposed to several different chemical challenges over 12 weeks of evolution, and whole-genome resequenced. Weekly forced outcrossing resulted in an average between adjacent-gene per cell division recombination rate of ∼0.0008. Despite attempts to force weekly sex, roughly half of our populations evolved cheaters and appear to be evolving asexually. Focusing on seven chemical stressors and 55 total evolved populations that remained sexual we observed large fitness gains and highly repeatable patterns of genome-wide haplotype change within chemical challenges, with limited levels of repeatability across chemical treatments. Adaptation appears highly polygenic with almost the entire genome showing significant and consistent patterns of haplotype change with little evidence for long-range linkage disequilibrium in a subset of populations for which we sequenced haploid clones. That is, almost the entire genome is under selection or drafting with selected sites. At any given locus adaptation was almost always dominated by one of the 18 founder's alleles, with that allele varying spatially and between treatments, suggesting that selection acts primarily on rare variants private to a founder or haplotype blocks harboring multiple mutations.
Keywords: complex traits; evolve and resequence; experimental evolution; yeast.
© The Author(s) 2022. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures



Similar articles
-
Standing genetic variation drives repeatable experimental evolution in outcrossing populations of Saccharomyces cerevisiae.Mol Biol Evol. 2014 Dec;31(12):3228-39. doi: 10.1093/molbev/msu256. Epub 2014 Aug 28. Mol Biol Evol. 2014. PMID: 25172959 Free PMC article.
-
Recombination Alters the Dynamics of Adaptation on Standing Variation in Laboratory Yeast Populations.Mol Biol Evol. 2018 Jan 1;35(1):180-201. doi: 10.1093/molbev/msx278. Mol Biol Evol. 2018. PMID: 29069452 Free PMC article.
-
Two Synthetic 18-Way Outcrossed Populations of Diploid Budding Yeast with Utility for Complex Trait Dissection.Genetics. 2020 Jun;215(2):323-342. doi: 10.1534/genetics.120.303202. Epub 2020 Apr 2. Genetics. 2020. PMID: 32241804 Free PMC article.
-
How does adaptation sweep through the genome? Insights from long-term selection experiments.Proc Biol Sci. 2012 Dec 22;279(1749):5029-38. doi: 10.1098/rspb.2012.0799. Epub 2012 Jul 25. Proc Biol Sci. 2012. PMID: 22833271 Free PMC article. Review.
-
Revisiting Mortimer's Genome Renewal Hypothesis: heterozygosity, homothallism, and the potential for adaptation in yeast.Adv Exp Med Biol. 2014;781:37-48. doi: 10.1007/978-94-007-7347-9_3. Adv Exp Med Biol. 2014. PMID: 24277294 Free PMC article. Review.
Cited by
-
The Dynamics of Adaptation to Stress from Standing Genetic Variation and de novo Mutations.Mol Biol Evol. 2022 Nov 3;39(11):msac242. doi: 10.1093/molbev/msac242. Mol Biol Evol. 2022. PMID: 36334099 Free PMC article.
-
MotifbreakR v2: extended capability and database integration.ArXiv [Preprint]. 2024 Jul 3:arXiv:2407.03441v1. ArXiv. 2024. PMID: 39010878 Free PMC article. Preprint.
-
Crossing design shapes patterns of genetic variation in synthetic recombinant populations of Saccharomyces cerevisiae.Sci Rep. 2021 Oct 1;11(1):19551. doi: 10.1038/s41598-021-99026-0. Sci Rep. 2021. PMID: 34599243 Free PMC article.
-
motifbreakR v2: expanded variant analysis including indels and integrated evidence from transcription factor binding databases.Bioinform Adv. 2024 Oct 23;4(1):vbae162. doi: 10.1093/bioadv/vbae162. eCollection 2024. Bioinform Adv. 2024. PMID: 39474623 Free PMC article.
References
-
- Barghi N, Hermisson J, Schlötterer C. 2020. Polygenic adaptation: a unifying framework to understand positive selection. Nat Rev Genet. 21:769–781. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

