Dynamic regulation of Zn(II) sequestration by calgranulin C
- PMID: 36367084
- PMCID: PMC9650546
- DOI: 10.1002/pro.4403
Dynamic regulation of Zn(II) sequestration by calgranulin C
Abstract
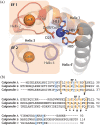
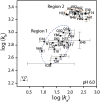
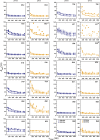
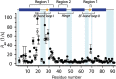
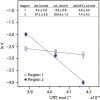
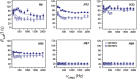
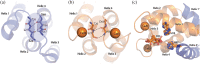
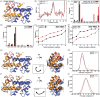
Calgranulin C performs antimicrobial activity in the human immune response by sequestering Zn(II). This biological function is afforded with the aid of two structurally distinct Ca(II)-binding EF hand motifs, wherein one of which bears an unusual amino acid sequence. Here, we utilize solution state NMR relaxation measurements to investigate the mechanism of Ca(II)-modulated enhancement of Zn(II) sequestration by calgranulin C. Using C13 /N15 CPMG dispersion experiments we have measured pH-dependent major and minor state populations exchanging on micro-to-millisecond timescale. This conformational exchange takes place exclusively in the Ca(II)-bound state and can be mapped to residues located in the EF-I loop and the linker between the tandem EF hands. Molecular dynamics (MD) simulations spanning nano-to-microsecond timescale offer insights into the role of pH-dependent electrostatic interactions in EF-hand dynamics. Our results suggest a pH-regulated dynamic equilibrium of conformations that explore a range of "closed" and partially "open" sidechain configurations within the Zn(II) binding site. We propose a novel mechanism by which Ca(II) binding to a non-canonical EF loop regulates its flexibility and tunes the antimicrobial activity of calgranulin C.
Keywords: Ca(II) binding proteins; NMR spectroscopy; molecular dynamics simulations; zinc sequestration.
© 2022 The Protein Society.
Figures

Similar articles
-
Calcium mediated static and dynamic allostery in S100A12: Implications for target recognition by S100 proteins.Protein Sci. 2024 Apr;33(4):e4955. doi: 10.1002/pro.4955. Protein Sci. 2024. PMID: 38501487 Free PMC article.
-
Molecular Basis of Ca(II)-Induced Tetramerization and Transition-Metal Sequestration in Human Calprotectin.J Am Chem Soc. 2021 Nov 3;143(43):18073-18090. doi: 10.1021/jacs.1c06402. Epub 2021 Oct 26. J Am Chem Soc. 2021. PMID: 34699194 Free PMC article.
-
Avian MRP126 Restricts Microbial Growth through Ca(II)-Dependent Zn(II) Sequestration.Biochemistry. 2020 Feb 18;59(6):802-817. doi: 10.1021/acs.biochem.9b01012. Epub 2020 Jan 21. Biochemistry. 2020. PMID: 31886651 Free PMC article.
-
Enzyme dynamics from NMR spectroscopy.Acc Chem Res. 2015 Feb 17;48(2):457-65. doi: 10.1021/ar500340a. Epub 2015 Jan 9. Acc Chem Res. 2015. PMID: 25574774 Free PMC article. Review.
-
Structural basis for diversity of the EF-hand calcium-binding proteins.J Mol Biol. 2006 Jun 9;359(3):509-25. doi: 10.1016/j.jmb.2006.03.066. Epub 2006 Apr 21. J Mol Biol. 2006. PMID: 16678204 Review.
Cited by
-
Calcium mediated static and dynamic allostery in S100A12: Implications for target recognition by S100 proteins.Protein Sci. 2024 Apr;33(4):e4955. doi: 10.1002/pro.4955. Protein Sci. 2024. PMID: 38501487 Free PMC article.
References
-
- Weinberg ED. Nutritional immunity: Host's attempt to withhold iron from microbial invaders. JAMA. 1975;231:39–41. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

