Loss of CD24 promotes radiation‑ and chemo‑resistance by inducing stemness properties associated with a hybrid E/M state in breast cancer cells
- PMID: 36367190
- PMCID: PMC9685273
- DOI: 10.3892/or.2022.8441
Loss of CD24 promotes radiation‑ and chemo‑resistance by inducing stemness properties associated with a hybrid E/M state in breast cancer cells
Abstract
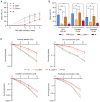
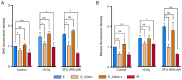
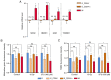
Cancer stem cells (CSCs) serve an essential role in failure of conventional antitumor therapy. In breast cancer, CD24‑/low/CD44+ phenotype and high aldehyde dehydrogenase activity are associated with CSC subtypes. Furthermore, CD24‑/low/CD44+ pattern is also characteristic of mesenchymal cells generated by epithelial‑mesenchymal transition (EMT). CD24 is a surface marker expressed in numerous types of tumor, however, its biological functions and role in cancer progression and treatment resistance remain poorly documented. Loss of CD24 expression in breast cancer cells is associated with radiation resistance and control of oxidative stress. Reactive oxygen species (ROS) mediate the effects of anticancer drugs as well as ionizing radiation; therefore, the present study investigated if CD24 mediates radiation‑ and chemo‑resistance of breast cancer cells. Using a HMLE breast cancer cell model, CD24 expression has been artificially modulated and it was observed that loss of CD24 expression induced stemness properties associated with acquisition of a hybrid E/M phenotype. CD24‑/low cells were more radiation‑ and chemo‑resistant than CD24+ cells. The resistance was associated with lower levels of ROS; CD24 controlled ROS levels via regulation of mitochondrial function independently of antioxidant activity. Together, these results suggested a key role of CD24 in de‑differentiation of breast cancer cells and promoting acquisition of therapeutic resistance properties.
Keywords: CD24; breast cancer; cancer stem cells; epithelial-mesenchymal transition; resistance.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
ALDH1A3 is the switch that determines the balance of ALDH+ and CD24-CD44+ cancer stem cells, EMT-MET, and glucose metabolism in breast cancer.Oncogene. 2024 Oct;43(43):3151-3169. doi: 10.1038/s41388-024-03156-4. Epub 2024 Sep 9. Oncogene. 2024. PMID: 39251846 Free PMC article.
-
Inhibition of Cdk2 kinase activity selectively targets the CD44⁺/CD24⁻/Low stem-like subpopulation and restores chemosensitivity of SUM149PT triple-negative breast cancer cells.Int J Oncol. 2014 Sep;45(3):1193-9. doi: 10.3892/ijo.2014.2523. Epub 2014 Jun 25. Int J Oncol. 2014. PMID: 24970653 Free PMC article.
-
CD44 and CD24 coordinate the reprogramming of nasopharyngeal carcinoma cells towards a cancer stem cell phenotype through STAT3 activation.Oncotarget. 2016 Sep 6;7(36):58351-58366. doi: 10.18632/oncotarget.11113. Oncotarget. 2016. PMID: 27521216 Free PMC article.
-
Breast cancer stem cells and intrinsic subtypes: controversies rage on.Curr Stem Cell Res Ther. 2009 Jan;4(1):50-60. doi: 10.2174/157488809787169110. Curr Stem Cell Res Ther. 2009. PMID: 19149630 Review.
-
Epithelial mesenchymal transition traits in human breast cancer cell lines parallel the CD44(hi/)CD24 (lo/-) stem cell phenotype in human breast cancer.J Mammary Gland Biol Neoplasia. 2010 Jun;15(2):235-52. doi: 10.1007/s10911-010-9175-z. Epub 2010 Jun 4. J Mammary Gland Biol Neoplasia. 2010. PMID: 20521089 Review.
Cited by
-
From mechanism to therapy: the journey of CD24 in cancer.Front Immunol. 2024 May 31;15:1401528. doi: 10.3389/fimmu.2024.1401528. eCollection 2024. Front Immunol. 2024. PMID: 38881902 Free PMC article. Review.
-
Identification of cell adhesion-related subtypes and construction of risk model to predict breast cancer prognostic and immunological properties.World J Surg Oncol. 2025 Apr 22;23(1):152. doi: 10.1186/s12957-025-03802-5. World J Surg Oncol. 2025. PMID: 40264099 Free PMC article.
-
Re-Sensitizing Cancer Stem Cells to Conventional Chemotherapy Agents.Int J Mol Sci. 2023 Jan 20;24(3):2122. doi: 10.3390/ijms24032122. Int J Mol Sci. 2023. PMID: 36768445 Free PMC article. Review.
-
Cancer stem cells: advances in knowledge and implications for cancer therapy.Signal Transduct Target Ther. 2024 Jul 5;9(1):170. doi: 10.1038/s41392-024-01851-y. Signal Transduct Target Ther. 2024. PMID: 38965243 Free PMC article. Review.
-
Development and Characterization of Three Novel FGFR Inhibitor Resistant Cervical Cancer Cell Lines to Help Drive Cervical Cancer Research.Int J Mol Sci. 2025 Feb 20;26(5):1799. doi: 10.3390/ijms26051799. Int J Mol Sci. 2025. PMID: 40076427 Free PMC article.
References
-
- Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

