Retrotransposon insertions associated with risk of neurologic and psychiatric diseases
- PMID: 36367221
- PMCID: PMC9827563
- DOI: 10.15252/embr.202255197
Retrotransposon insertions associated with risk of neurologic and psychiatric diseases
Abstract
Transposable elements (TEs) are active in neuronal cells raising the question whether TE insertions contribute to risk of neuropsychiatric disease. While genome-wide association studies (GWAS) serve as a tool to discover genetic loci associated with neuropsychiatric diseases, unfortunately GWAS do not directly detect structural variants such as TEs. To examine the role of TEs in psychiatric and neurologic disease, we evaluated 17,000 polymorphic TEs and find 76 are in linkage disequilibrium with disease haplotypes (P < 10-6 ) defined by GWAS. From these 76 polymorphic TEs, we identify potentially causal candidates based on having insertions in genomic regions of regulatory chromatin and on having associations with altered gene expression in brain tissues. We show that lead candidate insertions have regulatory effects on gene expression in human neural stem cells altering the activity of a minimal promoter. Taken together, we identify 10 polymorphic TE insertions that are potential candidates on par with other variants for having a causal role in neurologic and psychiatric disorders.
Keywords: Alu; GWAS; neurologic disease; psychiatric disease; transposable elements.
Published 2022. This article is a U.S. Government work and is in the public domain in the USA.
Figures
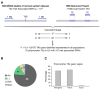
Polymorphic TEs within 1 Mb of a TAS were evaluated for LD with the TAS.
Classification of TEs in LD with TASs.
Genic location of TEs in LD with TASs.
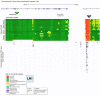
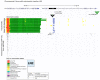

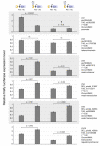
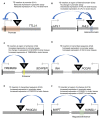
Locus number 1 (LN1) is located in promoter chromatin of TTLL4 and may disrupt promoter function.
LN2 is within an island of heterochromatin and may alter chromatin interactions to increase expression of UFL1 and reduce FHL5 expression.
LN3 is positioned within enhancer chromatin and may increase expression of TMEM266 by contributing activator sequence.
LN4 is located in transcribed sequence and may increase expression of INA by contributing activator sequence.
LN5 is located in transcribed sequence and may increase expression of PIK3C2A by contributing activator sequence.
LN16 is positioned in enhancer chromatin and may increase expression of MAPT and KANSL1 by contributing activator sequence.
References
-
- Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DA006227/DA/NIDA NIH HHS/United States
- R01 MH101782/MH/NIMH NIH HHS/United States
- R01 MH101810/MH/NIMH NIH HHS/United States
- R01 MH101819/MH/NIMH NIH HHS/United States
- R01 MH090936/MH/NIMH NIH HHS/United States
- R01 MH090951/MH/NIMH NIH HHS/United States
- R01 MH101820/MH/NIMH NIH HHS/United States
- R01 MH101822/MH/NIMH NIH HHS/United States
- R01 MH101825/MH/NIMH NIH HHS/United States
- R01 MH090948/MH/NIMH NIH HHS/United States
- R01 MH090941/MH/NIMH NIH HHS/United States
- HHSN261200800001C/RC/CCR NIH HHS/United States
- R01 MH090937/MH/NIMH NIH HHS/United States
- HHSN268201000029C/HL/NHLBI NIH HHS/United States
- HHSN261200800001E/CA/NCI NIH HHS/United States
- R01 MH101814/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials

