Inhibition of a malaria host-pathogen interaction by a computationally designed inhibitor
- PMID: 36367441
- PMCID: PMC9793980
- DOI: 10.1002/pro.4507
Inhibition of a malaria host-pathogen interaction by a computationally designed inhibitor
Abstract
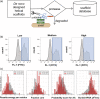
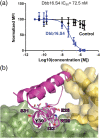
Malaria is a substantial global health burden with 229 million cases in 2019 and 450,000 deaths annually. Plasmodium vivax is the most widespread malaria-causing parasite putting 2.5 billion people at risk of infection. P. vivax has a dormant liver stage and therefore can exist for long periods undetected. Its blood-stage can cause severe reactions and hospitalization. Few treatment and detection options are available for this pathogen. A unique characteristic of P. vivax is that it depends on the Duffy antigen/receptor for chemokines (DARC) on the surface of host red blood cells for invasion. P. vivax employs the Duffy binding protein (DBP) to bind to DARC. We first de novo designed a three helical bundle scaffolding database which was screened via protease digestions for stability. Protease-resistant scaffolds highlighted thresholds for stability, which we utilized for selecting DARC mimetics that we subsequentially designed through grafting and redesign of these scaffolds. The optimized design small helical protein disrupts the DBP:DARC interaction. The inhibitor blocks the receptor binding site on DBP and thus forms a strong foundation for a therapeutic that will inhibit reticulocyte infection and prevent the pathogenesis of P. vivax malaria.
Keywords: Plasmodium vivax; protease selections; protein design; protein scaffolds; three-helical bundles.
© 2022 The Authors. Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- WHO . World malaria report 2017. Geneva: World Health Organization, 2017.
-
- Popovici J, Menard D. Challenges in antimalarial drug treatment for vivax malaria control. Trends Mol Med. 2015;21:776–788. - PubMed
-
- Krotoski WA. The hypnozoite and malarial relapse. Prog Clin Parasitol. 1989;1:1–19. - PubMed
-
- Adams JH, Hudson DE, Torii M, et al. The Duffy receptor family of Plasmodium knowlesi is located within the micronemes of invasive malaria merozoites. Cell. 1990;63:141–153. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

