MAGE-A3 regulates tumor stemness in gastric cancer through the PI3K/AKT pathway
- PMID: 36367777
- PMCID: PMC9792200
- DOI: 10.18632/aging.204373
MAGE-A3 regulates tumor stemness in gastric cancer through the PI3K/AKT pathway
Abstract
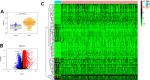
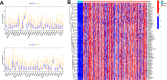
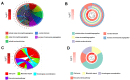
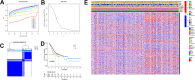
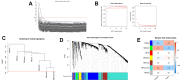
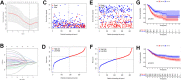
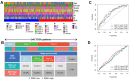
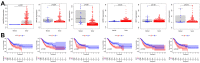
Gastric cancer remains a malignant disease of the digestive tract with high mortality and morbidity worldwide. However, due to its complex pathological mechanisms and lack of effective clinical therapies, the survival rate of patients after receiving treatment is not satisfactory. A increasing number of studies have focused on cancer stem cells and their regulatory properties. In this study, we first constructed a co-expression network based on the WGCNA algorithm to identify modules with different degrees of association with tumor stemness indices. After selecting the most positively correlated modules of the stemness index, we performed a consensus clustering analysis on gastric cancer samples and constructed the co-expression network again. We then selected the modules of interest and applied univariate COX regression analysis to the genes in this module for preliminary screening. The results of the screening were then used in LASSO regression analysis to construct a risk prognostic model and subsequently a sixteen-gene model was obtained. Finally, after verifying the accuracy of the module and screening for risk genes, we identified MAGE-A3 as the final study subject. We then performed in vivo and in vitro experiments to verify its effect on tumor stemness and tumour proliferation. Our data supports that MAGE-A3 is a tumor stemness regulator and a potent prognostic biomarker which can help the prediction and treatment of gastric cancer patients.
Keywords: MAGE-A3; WGCNA; gastric cancer; mRNAsi.
Conflict of interest statement
Figures



Similar articles
-
Identification of stemness index-related long noncoding RNA SNHG12 in human bladder cancer based on WGCNA.Mol Cell Probes. 2022 Dec;66:101867. doi: 10.1016/j.mcp.2022.101867. Epub 2022 Sep 29. Mol Cell Probes. 2022. PMID: 36183925
-
Melanoma associated antigen (MAGE)-A3 promotes cell proliferation and chemotherapeutic drug resistance in gastric cancer.Cell Oncol (Dordr). 2016 Apr;39(2):175-86. doi: 10.1007/s13402-015-0261-5. Epub 2016 Feb 11. Cell Oncol (Dordr). 2016. PMID: 26868260
-
[Dysregulation of MAD2L1/CAMK2A/PTTG1 Gene Cluster Maintains the Stemness Characteristics of Uterine Corpus Endometrial Carcinoma].Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2021 Oct;43(5):685-695. doi: 10.3881/j.issn.1000-503X.13271. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2021. PMID: 34728029 Chinese.
-
Identification of Glioma Cancer Stem Cell Characteristics Based on Weighted Gene Prognosis Module Co-Expression Network Analysis of Transcriptome Data Stemness Indices.J Mol Neurosci. 2020 Oct;70(10):1512-1520. doi: 10.1007/s12031-020-01590-z. Epub 2020 May 26. J Mol Neurosci. 2020. PMID: 32451841
-
LETM1 is a potential biomarker that predicts poor prognosis in gastric adenocarcinoma.Exp Mol Pathol. 2020 Feb;112:104333. doi: 10.1016/j.yexmp.2019.104333. Epub 2019 Nov 6. Exp Mol Pathol. 2020. PMID: 31705880
Cited by
-
NRP1 regulates autophagy and proliferation of gastric cancer through Wnt/β-catenin signaling pathway.Aging (Albany NY). 2023 Mar 7;15(17):8613-8629. doi: 10.18632/aging.204560. Epub 2023 Mar 7. Aging (Albany NY). 2023. PMID: 37702613 Free PMC article.
-
Apigenin promotes apoptosis of 4T1 cells through PI3K/AKT/Nrf2 pathway and improves tumor immune microenvironment in vivo.Toxicol Res (Camb). 2024 Jan 25;13(1):tfae011. doi: 10.1093/toxres/tfae011. eCollection 2024 Feb. Toxicol Res (Camb). 2024. PMID: 38283821 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

