High-temporal resolution profiling reveals distinct immune trajectories following the first and second doses of COVID-19 mRNA vaccines
- PMID: 36367935
- PMCID: PMC9651857
- DOI: 10.1126/sciadv.abp9961
High-temporal resolution profiling reveals distinct immune trajectories following the first and second doses of COVID-19 mRNA vaccines
Abstract
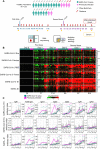
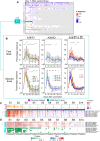
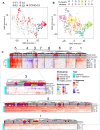
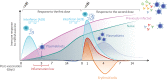
Knowledge of the mechanisms underpinning the development of protective immunity conferred by mRNA vaccines is fragmentary. Here, we investigated responses to coronavirus disease 2019 (COVID-19) mRNA vaccination via high-temporal resolution blood transcriptome profiling. The first vaccine dose elicited modest interferon and adaptive immune responses, which peaked on days 2 and 5, respectively. The second vaccine dose, in contrast, elicited sharp day 1 interferon, inflammation, and erythroid cell responses, followed by a day 5 plasmablast response. Both post-first and post-second dose interferon signatures were associated with the subsequent development of antibody responses. Yet, we observed distinct interferon response patterns after each of the doses that may reflect quantitative or qualitative differences in interferon induction. Distinct interferon response phenotypes were also observed in patients with COVID-19 and were associated with severity and differences in duration of intensive care. Together, this study also highlights the benefits of adopting high-frequency sampling protocols in profiling vaccine-elicited immune responses.
Figures





References
-
- Gaucher D., Therrien R., Kettaf N., Angermann B. R., Boucher G., Filali-Mouhim A., Moser J. M., Mehta R. S., Drake D. R., Castro E., Akondy R., Rinfret A., Yassine-Diab B., Said E. A., Chouikh Y., Cameron M. J., Clum R., Kelvin D., Somogyi R., Greller L. D., Balderas R. S., Wilkinson P., Pantaleo G., Tartaglia J., Haddad E. K., Sékaly R.-P., Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J. Exp. Med. 205, 3119–3131 (2008). - PMC - PubMed
-
- Querec T. D., Akondy R. S., Lee E. K., Cao W., Nakaya H. I., Teuwen D., Pirani A., Gernert K., Deng J., Marzolf B., Kennedy K., Wu H., Bennouna S., Oluoch H., Miller J., Vencio R. Z., Mulligan M., Aderem A., Ahmed R., Pulendran B., Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol. 10, 116–125 (2009). - PMC - PubMed
-
- Arunachalam P. S., Scott M. K. D., Hagan T., Li C., Feng Y., Wimmers F., Grigoryan L., Trisal M., Edara V. V., Lai L., Chang S. E., Feng A., Dhingra S., Shah M., Lee A. S., Chinthrajah S., Sindher S. B., Mallajosyula V., Gao F., Sigal N., Kowli S., Gupta S., Pellegrini K., Tharp G., Maysel-Auslender S., Hamilton S., Aoued H., Hrusovsky K., Roskey M., Bosinger S. E., Maecker H. T., Boyd S. D., Davis M. M., Utz P. J., Suthar M. S., Khatri P., Nadeau K. C., Pulendran B., Systems vaccinology of the BNT162b2 mRNA vaccine in humans. Nature 596, 410–416 (2021). - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

