Alignment and Analysis of a Disparately Acquired Multibatch Metabolomics Study of Maternal Pregnancy Samples
- PMID: 36367990
- PMCID: PMC11701891
- DOI: 10.1021/acs.jproteome.2c00371
Alignment and Analysis of a Disparately Acquired Multibatch Metabolomics Study of Maternal Pregnancy Samples
Abstract
Untargeted liquid chromatography-mass spectrometry metabolomics studies are typically performed under roughly identical experimental settings. Measurements acquired with different LC-MS protocols or following extended time intervals harbor significant variation in retention times and spectral abundances due to altered chromatographic, spectrometric, and other factors, raising many data analysis challenges. We developed a computational workflow for merging and harmonizing metabolomics data acquired under disparate LC-MS conditions. Plasma metabolite profiles were collected from two sets of maternal subjects three years apart using distinct instruments and LC-MS procedures. Metabolomics features were aligned using metabCombiner to generate lists of compounds detected across all experimental batches. We applied data set-specific normalization methods to remove interbatch and interexperimental variation in spectral intensities, enabling statistical analysis on the assembled data matrix. Bioinformatics analyses revealed large-scale metabolic changes in maternal plasma between the first and third trimesters of pregnancy and between maternal plasma and umbilical cord blood. We observed increases in steroid hormones and free fatty acids from the first trimester to term of gestation, along with decreases in amino acids coupled to increased levels in cord blood. This work demonstrates the viability of integrating nonidentically acquired LC-MS metabolomics data and its utility in unconventional metabolomics study designs.
Keywords: LC-HRMS; alignment; metabolomics; normalization; partial correlation; plasma; pregnancy.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

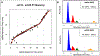
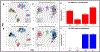
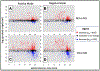

References
-
- Castillo S; Gopalacharyulu P; Yetukuri L; Orešič M Algorithms and Tools for the Preprocessing of LC–MS Metabolomics Data. Chemometrics and Intelligent Laboratory Systems 2011, 108 (1), 23–32.
-
- Computational Methods and Data Analysis for Metabolomics; Li S, Ed.; Methods in Molecular Biology; Humana Press: New York, NY, 2020.
-
- Jones OAH Illuminating the Dark Metabolome to Advance the Molecular Characterisation of Biological Systems. Metabolomics 2018, 14 (8), 101. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

