SpikeSeq: A rapid, cost efficient and simple method to identify SARS-CoV-2 variants of concern by Sanger sequencing part of the spike protein gene
- PMID: 36368344
- PMCID: PMC9642040
- DOI: 10.1016/j.jviromet.2022.114648
SpikeSeq: A rapid, cost efficient and simple method to identify SARS-CoV-2 variants of concern by Sanger sequencing part of the spike protein gene
Abstract
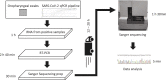
In 2020, the novel coronavirus, SARS-CoV-2, caused a pandemic, which is still raging at the time of writing this. Here, we present results from SpikeSeq, the first published Sanger sequencing-based method for the detection of Variants of Concern (VOC) and key mutations, using a 1 kb amplicon from the recognized ARTIC Network primers. The proposed setup relies entirely on materials and methods already in use in diagnostic RT-qPCR labs and on existing commercial infrastructure offering sequencing services. For data analysis, we provide an automated, open source, and browser-based mutation calling software (https://github.com/kblin/covid-spike-classification, https://ssi.biolib.com/covid-spike-classification). We validated the setup on 195 SARS-CoV-2 positive samples, and we were able to profile 85% of RT-qPCR positive samples, where the last 15% largely stemmed from samples with low viral count. We compared the SpikeSeq results to WGS results. SpikeSeq has been used as the primary variant identification tool on > 10.000 SARS-CoV-2 positive clinical samples during 2021. At approximately 4€ per sample in material cost, minimal hands-on time, little data handling, and a short turnaround time, the setup is simple enough to be implemented in any SARS-CoV-2 RT-qPCR diagnostic lab. Our protocol provides results that can be used to choose antibodies in a clinical setting and for the tracking and surveillance of all positive samples for new variants and known ones such as Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1) Delta (B.1.617.2), Omicron BA.1(B.1.1.529), BA.2, BA.4/5, BA.2.75.x, and many more, as of October 2022.
Keywords: BA.1; BA.2; BA.2.75.x; BA.4/5; COVID-19; CoV; Contact tracing; Coronavirus; Mutations; Omicron; Profiling; SARS-CoV-2; Sequencing; Spike protein; Surveillance; Variants of concern.
Copyright © 2022 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Andersen, C.Ø. et al. A major outbreak of COVID-19 at a residential care home. Dan. Med. J. 68(10):A03210227, 10 (2021). - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

