Genome-wide SNPs in the spiny lobster Panulirus homarus reveal a hybrid origin for its subspecies
- PMID: 36368918
- PMCID: PMC9652991
- DOI: 10.1186/s12864-022-08984-w
Genome-wide SNPs in the spiny lobster Panulirus homarus reveal a hybrid origin for its subspecies
Abstract
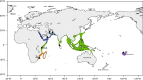
Background: Evolutionary divergence and speciation often occur at a slower rate in the marine realm due to the higher potential for long-distance reproductive interaction through larval dispersal. One common evolutionary pattern in the Indo-Pacific, is divergence of populations and species at the peripheries of widely-distributed organisms. However, the evolutionary and demographic histories of such divergence are yet to be well understood. Here we address these issues by coupling genome-wide SNP data with mitochondrial DNA sequences to test the patterns of genetic divergence and possible secondary contact among geographically distant populations of the highly valuable spiny lobster Panulirus homarus species complex, distributed widely through the Indo-Pacific, from South Africa to the Marquesas Islands.
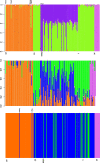
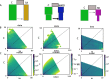
Result: After stringent filtering, 2020 SNPs were used for population genetic and demographic analyses, revealing strong regional structure (FST = 0.148, P < 0001), superficially in accordance with previous analyses. However, detailed demographic analyses supported a much more complex evolutionary history of these populations, including a hybrid origin of a North-West Indian Ocean (NWIO) population, which has previously been discriminated morphologically, but not genetically. The best-supported demographic models suggested that the current genetic relationships among populations were due to a complex series of past divergences followed by asymmetric migration in more recent times.
Conclusion: Overall, this study suggests that alternating periods of marine divergence and gene flow have driven the current genetic patterns observed in this lobster and may help explain the observed wider patterns of marine species diversity in the Indo-Pacific.
Keywords: Admixture; Demography; Genome-wide SNPs; Panulirus homarus complex.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no potential competing interest of interest.
Figures


References
-
- Bernardi G, Bucciarelli G, Costagliola D, Robertson DR, Heiser JB. Evolution of coral reef fish Thalassoma spp. (Labridae). 1. Molecular phylogeny and biogeography. Mar Biol. 2004;144:369–375. doi: 10.1007/s00227-003-1199-0. - DOI
-
- Ahti PA, Coleman RR, Dibattista JD, Berumen ML, Rocha LA, Bowen BW. Phylogeography of indo-Pacific reef fishes: sister wrasses Coris gaimard and C. cuvieri in the Red Sea, Indian Ocean and Pacific Ocean. J Biogeogr. 2016;43(6):1103–1115. doi: 10.1111/JBI.12712. - DOI
-
- Iacchei M, Gaither MR, Bowen BW, Toonen RJ. Testing dispersal limits in the sea: range-wide phylogeography of the pronghorn spiny lobster Panulirus penicillatus. J J Biogeogr. 2016;43(5):1032–1044. doi: 10.1111/jbi.12689. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

