Genetic regulation of serum IgA levels and susceptibility to common immune, infectious, kidney, and cardio-metabolic traits
- PMID: 36369178
- PMCID: PMC9651905
- DOI: 10.1038/s41467-022-34456-6
Genetic regulation of serum IgA levels and susceptibility to common immune, infectious, kidney, and cardio-metabolic traits
Erratum in
-
Author Correction: Genetic regulation of serum IgA levels and susceptibility to common immune, infectious, kidney, and cardio-metabolic traits.Nat Commun. 2023 Feb 6;14(1):655. doi: 10.1038/s41467-023-36340-3. Nat Commun. 2023. PMID: 36746961 Free PMC article. No abstract available.
Abstract
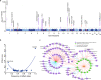
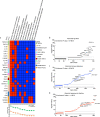
Immunoglobulin A (IgA) mediates mucosal responses to food antigens and the intestinal microbiome and is involved in susceptibility to mucosal pathogens, celiac disease, inflammatory bowel disease, and IgA nephropathy. We performed a genome-wide association study of serum IgA levels in 41,263 individuals of diverse ancestries and identified 20 genome-wide significant loci, including 9 known and 11 novel loci. Co-localization analyses with expression QTLs prioritized candidate genes for 14 of 20 significant loci. Most loci encoded genes that produced immune defects and IgA abnormalities when genetically manipulated in mice. We also observed positive genetic correlations of serum IgA levels with IgA nephropathy, type 2 diabetes, and body mass index, and negative correlations with celiac disease, inflammatory bowel disease, and several infections. Mendelian randomization supported elevated serum IgA as a causal factor in IgA nephropathy. African ancestry was consistently associated with higher serum IgA levels and greater frequency of IgA-increasing alleles compared to other ancestries. Our findings provide novel insights into the genetic regulation of IgA levels and its potential role in human disease.
© 2022. The Author(s).
Conflict of interest statement
Dr. Kiryluk has served on an advisory board for Goldfinch Bio and Gilead Sciences. Dr. Gharavi has served 1006 on an advisory board for Novartis, Travere and Natera and receives research grant funding from the Renal 1007 Research Institute and Natera. Dr. Moncrieffe is presently employed by Janssen Pharmaceutical Companies 37 1008 of Johnson & Johnson. Dr. Eitner is currently employed by Bayer AG. Drs. Julian and Novak are co-founders, 1009 co-owners of, and consultants for Reliant Glycosciences, LLC and are co-inventors on US patent application 1010 14/318,082 (assigned to UAB Research Foundation). The other authors declare no competing interests.
Figures




References
-
- Woof JM, Kerr MA. The function of immunoglobulin A in immunity. J. Pathol. 2006;208:270–282. - PubMed
-
- Woof JM, Russell MW. Structure and function relationships in IgA. Mucosal Immunol. 2011;4:590–597. - PubMed
-
- Corthesy B. Role of secretory IgA in infection and maintenance of homeostasis. Autoimmun. Rev. 2013;12:661–665. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 HG011172/HG/NHGRI NIH HHS/United States
- 75N92020D00001/HL/NHLBI NIH HHS/United States
- K25 DK128563/DK/NIDDK NIH HHS/United States
- N01 HC095167/HL/NHLBI NIH HHS/United States
- R01 MD012467/MD/NIMHD NIH HHS/United States
- U01 HG008679/HG/NHGRI NIH HHS/United States
- U01 HG008666/HG/NHGRI NIH HHS/United States
- R01 DK082753/DK/NIDDK NIH HHS/United States
- 75N92020D00002/HL/NHLBI NIH HHS/United States
- HHSN268201500003C/HL/NHLBI NIH HHS/United States
- U01 HG008676/HG/NHGRI NIH HHS/United States
- 75N92020D00005/HL/NHLBI NIH HHS/United States
- N01 HC095161/HL/NHLBI NIH HHS/United States
- U01 AI152960/AI/NIAID NIH HHS/United States
- N01 HC095168/HL/NHLBI NIH HHS/United States
- U24 NS107267/NS/NINDS NIH HHS/United States
- KL2 TR002737/TR/NCATS NIH HHS/United States
- R01 DK105124/DK/NIDDK NIH HHS/United States
- UL1 TR001079/TR/NCATS NIH HHS/United States
- N02 HL064278/HL/NHLBI NIH HHS/United States
- N01 HC095169/HL/NHLBI NIH HHS/United States
- R01 LM006910/LM/NLM NIH HHS/United States
- U01 HG008657/HG/NHGRI NIH HHS/United States
- K01 DK106341/DK/NIDDK NIH HHS/United States
- UL1 TR002736/TR/NCATS NIH HHS/United States
- N01 HC095159/HL/NHLBI NIH HHS/United States
- R01 DK078244/DK/NIDDK NIH HHS/United States
- 75N92020D00003/HL/NHLBI NIH HHS/United States
- U01 HG008672/HG/NHGRI NIH HHS/United States
- U19 AG065169/AG/NIA NIH HHS/United States
- R01 NS040807/NS/NINDS NIH HHS/United States
- P30 DK063491/DK/NIDDK NIH HHS/United States
- U01 HG008684/HG/NHGRI NIH HHS/United States
- UL1 TR001420/TR/NCATS NIH HHS/United States
- 75N92020D00004/HL/NHLBI NIH HHS/United States
- N01 HC095163/HL/NHLBI NIH HHS/United States
- 75N92020D00007/HL/NHLBI NIH HHS/United States
- R01 NS029993/NS/NINDS NIH HHS/United States
- R01 AR063759/AR/NIAMS NIH HHS/United States
- U54 MD007593/MD/NIMHD NIH HHS/United States
- HHSN268201500003I/HL/NHLBI NIH HHS/United States
- R03 DK122194/DK/NIDDK NIH HHS/United States
- U01 HG008680/HG/NHGRI NIH HHS/United States
- UL1 TR000040/TR/NCATS NIH HHS/United States
- U01 HG008673/HG/NHGRI NIH HHS/United States
- N01 HC095166/HL/NHLBI NIH HHS/United States
- U01 HG008685/HG/NHGRI NIH HHS/United States
- N01 HC095162/HL/NHLBI NIH HHS/United States
- U01 HG006379/HG/NHGRI NIH HHS/United States
- 75N92020D00006/HL/NHLBI NIH HHS/United States
- RC2 DK116690/DK/NIDDK NIH HHS/United States
- UL1 TR001873/TR/NCATS NIH HHS/United States
- UL1 TR001881/TR/NCATS NIH HHS/United States
- U01 HG008664/HG/NHGRI NIH HHS/United States
- N01 HC095165/HL/NHLBI NIH HHS/United States
- N01 HC095164/HL/NHLBI NIH HHS/United States
- U01 HG008701/HG/NHGRI NIH HHS/United States
- R01 LM013061/LM/NLM NIH HHS/United States
- N01 HC095160/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

