Molecular identity of proprioceptor subtypes innervating different muscle groups in mice
- PMID: 36369193
- PMCID: PMC9652284
- DOI: 10.1038/s41467-022-34589-8
Molecular identity of proprioceptor subtypes innervating different muscle groups in mice
Abstract
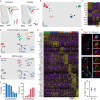
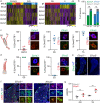
The precise execution of coordinated movements depends on proprioception, the sense of body position in space. However, the molecular underpinnings of proprioceptive neuron subtype identities are not fully understood. Here we used a single-cell transcriptomic approach to define mouse proprioceptor subtypes according to the identity of the muscle they innervate. We identified and validated molecular signatures associated with proprioceptors innervating back (Tox, Epha3), abdominal (C1ql2), and hindlimb (Gabrg1, Efna5) muscles. We also found that proprioceptor muscle identity precedes acquisition of receptor character and comprise programs controlling wiring specificity. These findings indicate that muscle-type identity is a fundamental aspect of proprioceptor subtype differentiation that is acquired during early development and includes molecular programs involved in the control of muscle target specificity.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Proske U, Gandevia SC. The proprioceptive senses: their roles in signaling body shape, body position and movement, and muscle force. Physiol. Rev. 2012;92:1651–1697. - PubMed
-
- Balaskas N, Ng D, Zampieri N. The positional logic of sensory-motor reflex circuit assembly. Neuroscience. 2020;450:142–150. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

