Stromal FOXF2 suppresses prostate cancer progression and metastasis by enhancing antitumor immunity
- PMID: 36369237
- PMCID: PMC9652358
- DOI: 10.1038/s41467-022-34665-z
Stromal FOXF2 suppresses prostate cancer progression and metastasis by enhancing antitumor immunity
Abstract
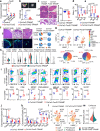
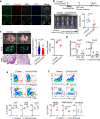
Cancer-associated fibroblasts (CAFs) mediate an immunosuppressive effect, but the underlying mechanism remains incompletely defined. Here we show that increasing prostatic stromal Foxf2 suppresses the growth and progression of both syngeneic and autochthonous mouse prostate cancer models in an immunocompetent context. Mechanistically, Foxf2 moderately attenuates the CAF phenotype and transcriptionally downregulates Cxcl5, which diminish the immunosuppressive myeloid cells and enhance T cell cytotoxicity. Increasing prostatic stromal Foxf2 sensitizes prostate cancer to the immune checkpoint blockade therapies. Augmenting lung stromal Foxf2 also mediates an immunosuppressive milieu and inhibits lung colonization of prostate cancer. FOXF2 is expressed higher in the stroma of human transition zone (TZ) than peripheral zone (PZ) prostate. The stromal FOXF2 expression level in primary prostate cancers inversely correlates with the Gleason grade. Our study establishes Foxf2 as a stromal transcription factor modulating the tumor immune microenvironment and potentially explains why cancers are relatively rare and indolent in the TZ prostate.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Wellenstein MD, de Visser KE. CanceR-cell-intrinsic mechanisms shaping the tumor immune landscape. Immunity. 2018;48:399–416. - PubMed
-
- Mhaidly R, Mechta-Grigoriou F. Fibroblast heterogeneity in tumor micro-environment: Role in immunosuppression and new therapies. Semin. Immunol. 2020;48:101417. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

